Team:Montgomery Cougars NJUSA/Project/MathematicalModel
From 2014hs.igem.org
(Difference between revisions)
(→Parameters) |
|||
| Line 20: | Line 20: | ||
<li>Doubling time of bacteria: | <li>Doubling time of bacteria: | ||
<li>The maximum rate at which DNA is transcribed in to mRNA in E.coli, as determined by the 2009 Beijing iGEM team is 4200nt/min. | <li>The maximum rate at which DNA is transcribed in to mRNA in E.coli, as determined by the 2009 Beijing iGEM team is 4200nt/min. | ||
| + | <li>The average speed of translation in E. coli is 2400Aa/min, as calculated by the 2009 Beijing iGEM team. | ||
</ul> | </ul> | ||
| + | |||
{{Montgomery_Cougars_NJUSA/Layout_End:Two_Column}} | {{Montgomery_Cougars_NJUSA/Layout_End:Two_Column}} | ||
Revision as of 18:44, 16 June 2014
Mathematical Model
Purpose
In order to obtain optimal results from each one of our experiments, the Montgomery iGEM team has undertaken the challenge of using differential calculus concepts to model the rates of growth of protein. We seek to determine the amount of proteins expressed by the bacteria's gene. The quantity of protein yielded by the bacteria is especially important due to the exacting amount of protein necessary to effectively alter enough sebum molecules and cut off P. acnes's nutrient source.
Defining our Model
Assumptions
- We are using the logistic growth equation to base our model on. We assume that bacteria follows logistical growth.
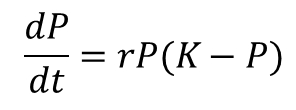
- The bacteria expresses the gene to produce the protein. We assume that the recombinant plasmid is effectively transformed into the bacteria.
Parameters
- Doubling time of bacteria:
- The maximum rate at which DNA is transcribed in to mRNA in E.coli, as determined by the 2009 Beijing iGEM team is 4200nt/min.
- The average speed of translation in E. coli is 2400Aa/min, as calculated by the 2009 Beijing iGEM team.
 "
"