Team:Montgomery Cougars NJUSA/Project/Labs
From 2014hs.igem.org
(→Our Experiences) |
(→Labs) |
||
| (63 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
* {{Montgomery_Cougars_NJUSA/Slider_Item | [[Image:DSC 7364.jpg|300px|link=]] | Micropipettes }} | * {{Montgomery_Cougars_NJUSA/Slider_Item | [[Image:DSC 7364.jpg|300px|link=]] | Micropipettes }} | ||
* {{Montgomery_Cougars_NJUSA/Slider_Item | [[Image:DSC 7381.jpg|300px|link=]] | Streaking a cell culture}} | * {{Montgomery_Cougars_NJUSA/Slider_Item | [[Image:DSC 7381.jpg|300px|link=]] | Streaking a cell culture}} | ||
| + | * {{Montgomery_Cougars_NJUSA/Slider_Item | [[Image:DSC 0201.jpg|300px|link=]] | Micropipetting }} | ||
| + | * {{Montgomery_Cougars_NJUSA/Slider_Item | [[Image:DSC 0002.jpg|300px|link=]] | Using Spin Columns }} | ||
| + | * {{Montgomery_Cougars_NJUSA/Slider_Item | [[Image:DSC 1003.jpg|300px|link=]] | Disposal of PlasticWare }} | ||
| + | * {{Montgomery_Cougars_NJUSA/Slider_Item | [[Image:DSC 0004.jpg|300px|link=]] | Prepping a Cell Culture }} | ||
| + | * {{Montgomery_Cougars_NJUSA/Slider_Item | [[Image:DSC 0005.jpg|300px|link=]] | Streaking on Agar }} | ||
| + | * {{Montgomery_Cougars_NJUSA/Slider_Item | [[Image:DSC 0006.jpg|300px|link=]] | Prepping a Cell Culture }} | ||
| + | * {{Montgomery_Cougars_NJUSA/Slider_Item | [[Image:DSC 79904.jpg|300px|link=]] | Safety First! }} | ||
{{Montgomery_Cougars_NJUSA/Slider_Close}} | {{Montgomery_Cougars_NJUSA/Slider_Close}} | ||
| Line 13: | Line 20: | ||
===Materials=== | ===Materials=== | ||
| + | |||
| + | [[Image:Lab2.jpg|right|200px|thumb|Preparing the Culture Tubes]] | ||
<ul> | <ul> | ||
| Line 52: | Line 61: | ||
===Our Experiences=== | ===Our Experiences=== | ||
| - | |||
| - | For growing up the cultures, although we completed the procedure correctly, we | + | [[Image:Lab1.jpg|left|200px|thumb|Preparing the Culture Tubes]] |
| + | |||
| + | This part of the lab was really the first time since our run-through pGLO lab that students had a chance to have first hand practice. During the Agar Streaking procedure, we encountered a significant setback to our already tight schedule. We had looked at the protocol for the 3A assembly kit, since parts were analogous, and streaked the bacteria onto Ampicillin/Kanamycin plates that were provided for us. We did not think that the parts we had ordered came with this resistance. So, we waited a full day to find that there was absolutely no growth. Thus, we had to repeat this all again on plain LB Agar. | ||
| + | |||
| + | For growing up the cultures, although we completed the procedure correctly, we did not end up using these cultures for our experiment. The one day waiting period was simply too long for our tight schedule. Our students talked with our advisor and we determined that the bacteria on the agar plate should be just as good as those that would grow within the next day. So, all of of subsequent experimental data comes from the growth on the original agar plates. | ||
==Miniprep== | ==Miniprep== | ||
| Line 83: | Line 95: | ||
===Our Experiences=== | ===Our Experiences=== | ||
| - | While we did not encounter many problems with this step directly, we did not use the standard cell cultures that were supposed to be used. We instead substituted for a faster mechanism which we did not see as significantly different. Due to our spin tube deficiency, we were forced to scale down our volumes to 1/10th their original values but kept all the proportions the same. In the end, we just ended up making very little of the purified DNA for the restriction digestion and ligation processes. We hope that our last-minute substitution plans do not interfere with the outcomes of our lab | + | |
| + | [[Image:Lab3.jpg|right|200px|thumb|Using the Centrifuge]] | ||
| + | |||
| + | While we did not encounter many problems with this step directly, we did not use the standard cell cultures that were supposed to be used. We instead substituted for a faster mechanism which we did not see as significantly different. Due to our spin tube deficiency, we were forced to scale down our volumes to 1/10th their original values but kept all the proportions the same. In the end, we just ended up making very little of the purified DNA for the restriction digestion and ligation processes. We hope that our last-minute substitution plans do not interfere with the outcomes of our lab. Other than that, we followed all of the standard protocol but skipped the optional but recommended steps. We ran out of time to do these. | ||
==Restriction Digest== | ==Restriction Digest== | ||
| Line 90: | Line 105: | ||
<ul> | <ul> | ||
<li>Ice and bucket/container</li> | <li>Ice and bucket/container</li> | ||
| - | |||
<li>Promoter (BBa_R0010) (Purified DNA, > 16ng/ul)</li> | <li>Promoter (BBa_R0010) (Purified DNA, > 16ng/ul)</li> | ||
<li>RBS (BBa_B0034) (Purified DNA, > 16ng/ul)</li> | <li>RBS (BBa_B0034) (Purified DNA, > 16ng/ul)</li> | ||
| - | <li>Med-Chain Aldehyde Dehydrogenase ( | + | <li>Med-Chain Aldehyde Dehydrogenase (BBa_K398005)(Purified DNA, > 16ng/ul)</li> |
| - | <li>Med-Chain Alcohol Dehydrogenase ( | + | <li>Med-Chain Alcohol Dehydrogenase (BBa_K398006) (Purified DNA, > 16ng/ul)</li> |
<li>GFP (BBa_E0040)(Purified DNA, > 16ng/ul)</li> | <li>GFP (BBa_E0040)(Purified DNA, > 16ng/ul)</li> | ||
<li>Terminator (BBa_B0015) (Purified DNA, > 16ng/ul)</li> | <li>Terminator (BBa_B0015) (Purified DNA, > 16ng/ul)</li> | ||
| Line 100: | Line 114: | ||
<li>dH2O</li> | <li>dH2O</li> | ||
<li>NEB Buffer 2</li> | <li>NEB Buffer 2</li> | ||
| - | |||
<li>Restriction Enzymes: EcoRI, SpeI, XbaI, PstI</li> | <li>Restriction Enzymes: EcoRI, SpeI, XbaI, PstI</li> | ||
<li>Thermal cycler</li> | <li>Thermal cycler</li> | ||
| Line 107: | Line 120: | ||
=== Procedure === | === Procedure === | ||
<ol> | <ol> | ||
| - | <li>Keep all enzymes and buffers used on ice | + | <li>Keep all enzymes and buffers used on ice to prevent degredation</li> |
| - | <li>Thaw NEB Buffer 2 | + | <li>Thaw NEB Buffer 2 at room temperature (We did not have BSA and instead used distilled water to fill volume. Collect solutions at the bottoms of the tubes.</li> |
| - | <li>Add 250ng of DNA to | + | <li>Add 250ng of DNA (from all 4 parts) to tubes. Add distilled water to the tubes for a total volume of 16ul in each tube</li> |
| - | + | <li>Add 2.5ul of NEB Buffer 2 to each tube.</li> | |
| - | + | <li>In the Promoter tube: Add 0.5ul of EcoRI, and 0.5ul of SpeI.</li> | |
| - | <li> | + | <li>In the RBS tube: Add 0.5ul of XbaI, and 0.5ul of SpeI.</li> |
| - | <li> | + | <li>In the Aldehyde Dehydrogenase tube: Add 0.5ul of XbaI, and 0.5ul of SpeI.</li> |
| - | <li>In the | + | <li>In the Alcohol Dehydrogenase tube: Add 0.5ul of XbaI, and 0.5ul of SpeI.</li> |
| - | <li>In the | + | <li>In the Terminator tube: Add 0.5ul of XbaI, and 0.5ul of PstI.</li> |
| - | <li>In the | + | <li>In the GFP tube: Add 0.5ul of XbaI, and 0.5ul of SpeI.</li> |
| + | <li>In the pSB1C3 tube: The pSB1C3 tube was already precut with EcoRI and PstI.</li> | ||
<li>The total volume in each tube should be approximately 20ul. Mix well by pipetting slowly up and down. Spin the samples briefly to collect all of the mixture to the bottom of the tube.</li> | <li>The total volume in each tube should be approximately 20ul. Mix well by pipetting slowly up and down. Spin the samples briefly to collect all of the mixture to the bottom of the tube.</li> | ||
<li>Incubate the restriction digests at 37°C for 30 minutes, then 80°C for 20 minutes. We use a thermal cycler with a heated lid.</li> | <li>Incubate the restriction digests at 37°C for 30 minutes, then 80°C for 20 minutes. We use a thermal cycler with a heated lid.</li> | ||
| - | |||
<li>Use ~2ul of the digest (25ng of DNA) for ligations.</li> | <li>Use ~2ul of the digest (25ng of DNA) for ligations.</li> | ||
</ol> | </ol> | ||
| - | + | ===Our Experience=== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | [[Image:Lab4.jpg|left|400px|thumb|R&D Leader Pipetting]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | It took us a while to figure out which restriction enzymes should be used to cut which parts; those difficulties and eventual solutions will be detailed in the space provided for the Ligation lab. The restriction digestion lab was completed smoothly, except for some issues with calculating appropriate molarity and volumes for components in the lab procedure. For example, we were not exactly sure how much distilled water to pipette into the reaction tube so we decided to pipette 6 ul of dH20 based on the protocol on the iGem website. Unfortunately, we may have used too little dH20, as it was difficult to retrieve the restriction digests for the ligation lab. | |
==Ligation== | ==Ligation== | ||
=== Materials === | === Materials === | ||
| + | |||
| + | <ol> | ||
| + | <li>Restriction digests produced from previous lab</li> | ||
| + | <li>T4 DNA ligase buffer</li> | ||
| + | <li>Rehydrated DNA from DNA Kit well plates</li> | ||
| + | <li>T4 DNA ligase</li> | ||
| + | <li>Thermal cycler</li> | ||
| + | </ol> | ||
=== Procedure === | === Procedure === | ||
<ol> | <ol> | ||
| - | <li>Add | + | <li>Add 3ul of promoter digested fragment.</li> |
| - | + | <li>Add 3ul of RBS digested fragment.</li> | |
| - | <li>Add | + | <li>Add 1 ul T4 DNA ligase buffer.</li> |
| - | <li>Add 1 ul T4 DNA ligase buffer. | + | <li>Add 0.5 ul T4 DNA ligase.</li> |
| - | <li>Add 0.5 ul T4 DNA ligase</li> | + | <li>Add water to 10 ul.</li> |
| - | <li>Add water to 10 ul</li> | + | |
<li>Ligate 16C/30 min, heat kill 80C/20 min</li> | <li>Ligate 16C/30 min, heat kill 80C/20 min</li> | ||
| - | <li>Transform with 1- | + | <li>Repeat steps 1-6 with the Aldehyde dehydrogenase, GFP, and terminator restricted digests.</li> |
| + | <li>Repeat steps 1-6 with the Alcohol dehydrogenase, GFP, and terminator restricted digests.</li> | ||
| + | <li>Repeat steps 1-6 with the resulting ligated parts and 2ul of digested plasmid backbone (Promoter/RBS and ORF/GFP/terminator and pSB1C3).</li> | ||
| + | <li>Transform with 1-2ul of product</li> | ||
</ol> | </ol> | ||
| + | |||
| + | The below diagram should further elucidate our procedure in creating the plasmid in interest. | ||
| + | |||
| + | [[Image:pink.jpg|center|700px|Ligation Diagram]] | ||
| + | |||
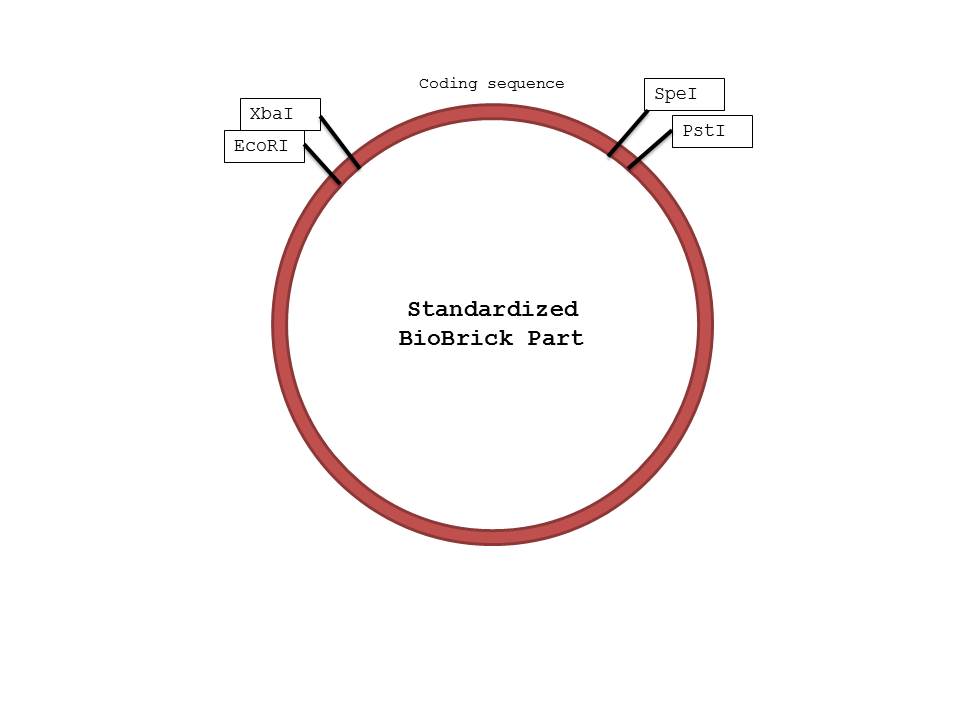
| + | [[Image:bb.jpg|center|600px|BioBrick]] | ||
===Our Experience=== | ===Our Experience=== | ||
| + | |||
| + | [[Image:Lab5.jpg|right|200px|thumb|Student preparing restriction tube]] | ||
| + | |||
| + | We were not surprised that when two parts are cut with the same enzyme, they could be successfully ligated together as their sticky ends were complementary. However, we were confused by the reason parts cut with XbaI and SpeI could also be ligated together, because that was how the mechanism seemed to work based on the protocol found on the iGEM website. When we researched further, we realized that both XbaI and SpeI cut to produce complementary sticky ends; only a few nucleotides upstream and downstream of the restriction site differed. By keeping the standard structure of Biobrick parts and the fact that parts cut with XbaI and SpeI could be ligated together in mind, we were able to design a procedure that would ensure proper parts were constructed in the order needed. (Please note that the ORF and the GFP genes could have been switched in the ligation process, as we ligated 3 parts at the same time, but in this case the order should not matter because they will both be transcribed and translated regardless) | ||
==Transformation== | ==Transformation== | ||
| Line 230: | Line 210: | ||
===Our Experiences=== | ===Our Experiences=== | ||
| + | |||
| + | [[Image:Lab6.jpg|left|200px|thumb|Plated out Transformations]] | ||
| + | |||
Our transformation procedure differed from the one listed on the registry. Due to our limited time, we opted to do only the absolutely necessary steps and combined practices seen from the pGLO lab. We did not include the RFP control or use antibiotics on our plates. The last time we used antibiotics, our parts failed to grow so we decided to run the risk of overgrowth versus nothing at all. Additionally, since we had extra ligated DNA, We inoculated 4 tubes of competent cells and plated all of them. We could not figure out why the experiment called for 20 and 200 microliters to plate so we plated all at 20. | Our transformation procedure differed from the one listed on the registry. Due to our limited time, we opted to do only the absolutely necessary steps and combined practices seen from the pGLO lab. We did not include the RFP control or use antibiotics on our plates. The last time we used antibiotics, our parts failed to grow so we decided to run the risk of overgrowth versus nothing at all. Additionally, since we had extra ligated DNA, We inoculated 4 tubes of competent cells and plated all of them. We could not figure out why the experiment called for 20 and 200 microliters to plate so we plated all at 20. | ||
{{Montgomery_Cougars_NJUSA/Layout_End:Two_Column}} | {{Montgomery_Cougars_NJUSA/Layout_End:Two_Column}} | ||
Latest revision as of 02:48, 21 June 2014
Labs
Contents |
Growing Cultures
Materials
- Agar Stab for Promotor BBa_R0010
- Agar Stab for RBS BBa_B0034
- Agar Stab for Protein Coding Domains BBa_K398006 and BBa_K398005
- Agar Stab for GFP BBa_E0040
- Agar Stab for Terminator BBa_B0015
- Inoculating Tubes
- LB Agar Plates
- Marker
- Inoculating Loops
- Snap Cap Tubes
- LB Broth
Procedure (Streaking Agar Plates)
- Clean the table and clear a lab space to have a contaminant-free culture
- Label each of the 4 plates with the appropriate part name, in this case, BBa_B0034, BBa_K398005, BBa_K398006, and BBa_E0040
- Use the inoculating loop to take a sample from each of the 4 agar stabs where the hole is visible
- Streak the loop with the bacteria sample onto the agar plate in zigzags, and once again 90 degrees rotated
- Repeat these steps for all 4 parts onto different agar plates
- Tape plates together and turn upside down so as to avoid condensation contamination
- Store in incubator at 37 degrees Celsius for 14-16 hours
- Agar plates, once incubated, can be stored in the fridge
Procedure (Growing Up Cultures)
- Clean table and clear a lab space to have a contaminant-free culture
- Take out the 4 plated agar samples from the incubator, there should be significant growth
- Using a pipette, put 5ml of LB nutrient broth into 4 snap tubes
- Use an inoculating loop to take colonies from each agar plate and swirl into a tube
- Close tubes and incubate for 14-16 hours
Our Experiences
This part of the lab was really the first time since our run-through pGLO lab that students had a chance to have first hand practice. During the Agar Streaking procedure, we encountered a significant setback to our already tight schedule. We had looked at the protocol for the 3A assembly kit, since parts were analogous, and streaked the bacteria onto Ampicillin/Kanamycin plates that were provided for us. We did not think that the parts we had ordered came with this resistance. So, we waited a full day to find that there was absolutely no growth. Thus, we had to repeat this all again on plain LB Agar.
For growing up the cultures, although we completed the procedure correctly, we did not end up using these cultures for our experiment. The one day waiting period was simply too long for our tight schedule. Our students talked with our advisor and we determined that the bacteria on the agar plate should be just as good as those that would grow within the next day. So, all of of subsequent experimental data comes from the growth on the original agar plates.
Miniprep
Materials
- Qiagen Miniprep Kit
- Cell Culture
- 1.6 ml Microcentrifuge tubes (2 per a miniprep)
- TE (1:10)
Procedure
- We opted not to use our grownup cell cultures. So we inoculated LB broth with the 4 respective parts and spun that culture in a centrifuge to pellet the cells, emptying the supernatant when complete
- Resuspend pelleted bacterial cells (all 4 samples) in 250 µl Buffer P1 and transfer to a microcentrifuge tube (we used just regular ones to accommodate our centrifuge)
- Add 250 μl Buffer P2 to the solution and gently mix. This allows for the lysis process to occur
- Add 350 μl Buffer N3 to each sample and gently mix.
- Centrifuge for 10 min at 13,000 rpm (~17,900 x g). In the end, a white pellet formed in each of the 4 samples we were miniprepping.
- Apply supernatants from step 4 to the QIAprep spin column by pipetting.
- Centrifuge for 30–60 and discard all the flow-through.
- Wash QIAprep spin column by adding 0.75 ml Buffer PE and centrifuging for 30–60 s.
- Discard the flow-through, and centrifuge for an additional 1 min to remove residual wash buffer.
- Place the QIAprep column in a clean 1.5 ml microcentrifuge tube. To elute DNA, add 50 μl Buffer EB (10 mM Tris·Cl, pH 8.5) or water to the center of each QIAprep spin column, let stand for 1 min, and centrifuge for 1 min.
Our Experiences
While we did not encounter many problems with this step directly, we did not use the standard cell cultures that were supposed to be used. We instead substituted for a faster mechanism which we did not see as significantly different. Due to our spin tube deficiency, we were forced to scale down our volumes to 1/10th their original values but kept all the proportions the same. In the end, we just ended up making very little of the purified DNA for the restriction digestion and ligation processes. We hope that our last-minute substitution plans do not interfere with the outcomes of our lab. Other than that, we followed all of the standard protocol but skipped the optional but recommended steps. We ran out of time to do these.
Restriction Digest
Materials
- Ice and bucket/container
- Promoter (BBa_R0010) (Purified DNA, > 16ng/ul)
- RBS (BBa_B0034) (Purified DNA, > 16ng/ul)
- Med-Chain Aldehyde Dehydrogenase (BBa_K398005)(Purified DNA, > 16ng/ul)
- Med-Chain Alcohol Dehydrogenase (BBa_K398006) (Purified DNA, > 16ng/ul)
- GFP (BBa_E0040)(Purified DNA, > 16ng/ul)
- Terminator (BBa_B0015) (Purified DNA, > 16ng/ul)
- Linearized plasmid backbone (pSB1C3)(25ng/ul)
- dH2O
- NEB Buffer 2
- Restriction Enzymes: EcoRI, SpeI, XbaI, PstI
- Thermal cycler
Procedure
- Keep all enzymes and buffers used on ice to prevent degredation
- Thaw NEB Buffer 2 at room temperature (We did not have BSA and instead used distilled water to fill volume. Collect solutions at the bottoms of the tubes.
- Add 250ng of DNA (from all 4 parts) to tubes. Add distilled water to the tubes for a total volume of 16ul in each tube
- Add 2.5ul of NEB Buffer 2 to each tube.
- In the Promoter tube: Add 0.5ul of EcoRI, and 0.5ul of SpeI.
- In the RBS tube: Add 0.5ul of XbaI, and 0.5ul of SpeI.
- In the Aldehyde Dehydrogenase tube: Add 0.5ul of XbaI, and 0.5ul of SpeI.
- In the Alcohol Dehydrogenase tube: Add 0.5ul of XbaI, and 0.5ul of SpeI.
- In the Terminator tube: Add 0.5ul of XbaI, and 0.5ul of PstI.
- In the GFP tube: Add 0.5ul of XbaI, and 0.5ul of SpeI.
- In the pSB1C3 tube: The pSB1C3 tube was already precut with EcoRI and PstI.
- The total volume in each tube should be approximately 20ul. Mix well by pipetting slowly up and down. Spin the samples briefly to collect all of the mixture to the bottom of the tube.
- Incubate the restriction digests at 37°C for 30 minutes, then 80°C for 20 minutes. We use a thermal cycler with a heated lid.
- Use ~2ul of the digest (25ng of DNA) for ligations.
Our Experience
It took us a while to figure out which restriction enzymes should be used to cut which parts; those difficulties and eventual solutions will be detailed in the space provided for the Ligation lab. The restriction digestion lab was completed smoothly, except for some issues with calculating appropriate molarity and volumes for components in the lab procedure. For example, we were not exactly sure how much distilled water to pipette into the reaction tube so we decided to pipette 6 ul of dH20 based on the protocol on the iGem website. Unfortunately, we may have used too little dH20, as it was difficult to retrieve the restriction digests for the ligation lab.
Ligation
Materials
- Restriction digests produced from previous lab
- T4 DNA ligase buffer
- Rehydrated DNA from DNA Kit well plates
- T4 DNA ligase
- Thermal cycler
Procedure
- Add 3ul of promoter digested fragment.
- Add 3ul of RBS digested fragment.
- Add 1 ul T4 DNA ligase buffer.
- Add 0.5 ul T4 DNA ligase.
- Add water to 10 ul.
- Ligate 16C/30 min, heat kill 80C/20 min
- Repeat steps 1-6 with the Aldehyde dehydrogenase, GFP, and terminator restricted digests.
- Repeat steps 1-6 with the Alcohol dehydrogenase, GFP, and terminator restricted digests.
- Repeat steps 1-6 with the resulting ligated parts and 2ul of digested plasmid backbone (Promoter/RBS and ORF/GFP/terminator and pSB1C3).
- Transform with 1-2ul of product
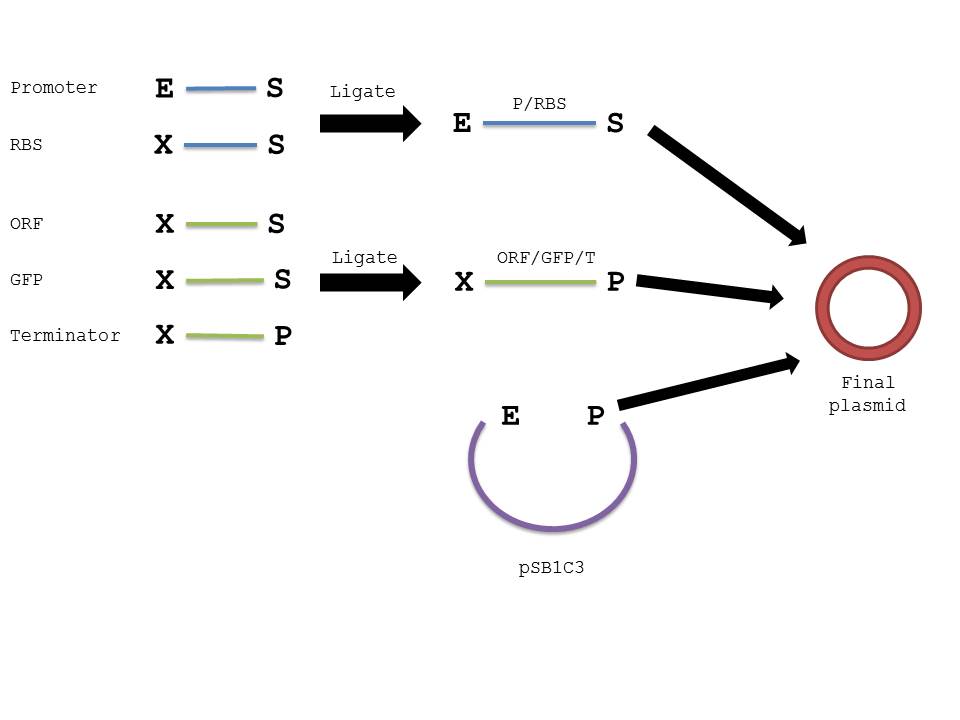
The below diagram should further elucidate our procedure in creating the plasmid in interest.
Our Experience
We were not surprised that when two parts are cut with the same enzyme, they could be successfully ligated together as their sticky ends were complementary. However, we were confused by the reason parts cut with XbaI and SpeI could also be ligated together, because that was how the mechanism seemed to work based on the protocol found on the iGEM website. When we researched further, we realized that both XbaI and SpeI cut to produce complementary sticky ends; only a few nucleotides upstream and downstream of the restriction site differed. By keeping the standard structure of Biobrick parts and the fact that parts cut with XbaI and SpeI could be ligated together in mind, we were able to design a procedure that would ensure proper parts were constructed in the order needed. (Please note that the ORF and the GFP genes could have been switched in the ligation process, as we ligated 3 parts at the same time, but in this case the order should not matter because they will both be transcribed and translated regardless)
Transformation
Materials
- Resuspended DNA
- Competent Cells
- Ice
- 2ml tube
- 42ºC water bath
- SOC media
- Agar Plates
- Glass beads
- 37ºC incubator
Procedure
- Add 50 µL of thawed competent cells into pre-chilled 2ml tube. We did not make these but rather took them from the BIORAD pGLO lab kit
- Add 1.5 µL of the resuspended DNA to the 2ml tube. Aspirate to mix, keeping the competent cells on ice.
- Close tubes and incubate the cells on ice for 30 minutes.
- Heat shock the cells by immersion in a pre-heated water bath at 42ºC for 60 seconds.
- Incubate the cells on ice for 5 minutes.
- Add 200 μl of SOC media
- Incubate the cells at 37ºC for 2 hours
- Label two petri dishes with LB agar.
- Plate 20 µl and 200 µl of the transformation onto the dishes, and spread with glass beads
- Incubate the plates at 37ºC for 12-14 hours, making sure the agar side of the plate is up.
Our Experiences
Our transformation procedure differed from the one listed on the registry. Due to our limited time, we opted to do only the absolutely necessary steps and combined practices seen from the pGLO lab. We did not include the RFP control or use antibiotics on our plates. The last time we used antibiotics, our parts failed to grow so we decided to run the risk of overgrowth versus nothing at all. Additionally, since we had extra ligated DNA, We inoculated 4 tubes of competent cells and plated all of them. We could not figure out why the experiment called for 20 and 200 microliters to plate so we plated all at 20.
 "
"