Team:Montgomery Cougars NJUSA/Project/Labs
From 2014hs.igem.org
(Difference between revisions)
(→Labs) |
(→Labs) |
||
| Line 5: | Line 5: | ||
== Growing Cultures == | == Growing Cultures == | ||
| + | |||
| + | ===Materials=== | ||
<ul> | <ul> | ||
Revision as of 19:07, 17 June 2014
Labs
Contents |
Growing Cultures
Materials
- Agar Stab for Promotor BBa_R0010
- Agar Stab for Terminator BBa_B0015
- Agar Stab for RBS BBa_B0034
- Agar Stab for GFP BBa_E0040
- Agar Stab for Protein Coding Domains BBa_K398006 and BBa_K398005
- Inoculating Tubes
- LB Agar Plates)
- Marker
- Inoculating Tubes
Miniprep
Materials
- Qiagen Miniprep Kit
- Cell Culture
- 1.6 ml Microcentrifuge tubes (2 per a miniprep)
- TE (1:10)
Procedure
- Spin the cell culture in a centrifuge to pellet the cells, empty the supernatant (media) into a waste collection container.
- Resuspend pelleted bacterial cells in 250 µl Buffer P1 (kept at 4 °C) and transfer to a microcentrifuge tube. No cell clumps should be visible after resuspension of the pellet. Important: Ensure that RNase A has been added to Buffer P1.
- Add 250 μl Buffer P2 and gently invert the tube 4–6 times to mix. Do not vortex, as this will result in shearing of genomic DNA. If necessary, continue inverting the tube until the solution becomes viscous and slightly clear. Do not allow the lysis reaction to proceed for more than 5 min.
- Add 350 μl Buffer N3 and invert the tube immediately and gently 4–6 times. To avoid localized precipitation, mix the solution gently but thoroughly, immediately after addition of Buffer N3. The solution should become cloudy.
- Centrifuge for 10 min at 13,000 rpm (~17,900 x g) in a table-top microcentrifuge. A white pellet will form.
- Apply the supernatants from step 4 to the QIAprep spin column by decanting or pipetting.
- Centrifuge for 30–60 s. Discard the flow-through.
- (Optional): Wash the QIAprep spin column by adding 0.5 ml Buffer PB and centrifuging for 30–60 s. Discard the flow-through.
- This step is necessary to remove trace nuclease activity when using endA+ strains such as the JM series, HB101 and its derivatives, or any wild-type strain, which have high levels of nuclease activity or high carbohydrate content. Host strains such as XL-1 Blue and DH5α™ do not require this additional wash step.
- Although they call this step optional, it does not really hurt your yield and you may think you are working with an endA- strain when in reality you are not. Again for this step, spinning for 60 seconds produces good results.
- Wash QIAprep spin column by adding 0.75 ml Buffer PE and centrifuging for 30–60 s.
- Spinning for 60 seconds produces good results.
- Discard the flow-through, and centrifuge for an additional 1 min to remove residual wash buffer.
- IMPORTANT: Residual wash buffer will not be completely removed unless the flow-through is discarded before this additional centrifugation. Residual ethanol from Buffer PE may inhibit subsequent enzymatic reactions. They are right about this.
- Place the QIAprep column in a clean 1.5 ml microcentrifuge tube. To elute DNA, add 50 μl Buffer EB (10 mM Tris·Cl, pH 8.5) or water to the center of each QIAprep spin column, let stand for 1 min, and centrifuge for 1 min.
Our Experience
Restriction Lab
Materials
- Ice and bucket/container
- (1) 8-tube strip, or (3) 0.6ml thin-walled tubes
- Part A (Purified DNA, > 16ng/ul)
- Part B (Purified DNA, > 16ng/ul)
- Linearized plasmid backbone (25ng/ul)
- dH2O
- NEB Buffer 2
- BSA
- Restriction Enzymes: EcoRI, SpeI, XbaI, PstI, DpnI
- Thermal cycler
Notes:
- You should keep all materials on ice.
- At iGEM HQ we use restriction enzymes from New England Biolabs
Procedure
- Keep all enzymes and buffers used on ice.
- Thaw NEB Buffer 2 and BSA in room temperature water. Mix by shaking the tubes, and flick/spin them to collect the liquid at the bottom of the tube.
- Add 250ng of DNA to the appropriately labelled tube. Add distilled water to the tubes for a total volume of 16ul in each tube. [Calculation example (with 25ng/ul as DNA sample concentration): 250ng ÷ 25ng/ul = 10ul of DNA sample 16ul (total volume) – 10ul (DNA sample) = 6ul of distilled water]
- Pipet 2.5ul of NEB Buffer 2 to each tube.
- Pipet 0.5ul of BSA to each tube.
- In the Part A tube: Add 0.5ul of EcoRI, and 0.5ul of SpeI.
- In the Part B tube: Add 0.5ul of XbaI, and 0.5ul of PstI.
- In the pSB1C3 tube: Add 0.5ul of EcoRI, 0.5ul of PstI, and 0.5ul of Dpn1.
- The total volume in each tube should be approximately 20ul. Mix well by pipetting slowly up and down. Spin the samples briefly to collect all of the mixture to the bottom of the tube.
- Incubate the restriction digests at 37°C for 30 minutes, then 80°C for 20 minutes. We use a thermal cycler with a heated lid.
- (Optional, but recommended) Run a portion of the digest on a gel (8ul, 100ng), to check that both plasmid backbone and part length are accurate.
- Use ~2ul of the digest (25ng of DNA) for ligations.
| Part A | Part B | Linearized Plasmid Backbone | |
|---|---|---|---|
| DNA | 250ng | 250ng | 250ng (10ul @ 25ng/ul) |
| dH2O | adjust to 16ul | adjust to 16ul | 16ul |
| NEB Buffer 2 | 2.5ul | 2.5ul | 2.5ul |
| NEB Buffer 2 | 0.5ul | 0.5ul | 0.5ul |
| Enzyme 1 | 0.5ul EcoRI | 0.5ul xBa1 | 0.5ul EcoRI |
| Enzyme 2 | 0.5ul SpeI | 0.5ul PstI | 0.5ul Pst1 |
| Enzyme 3 | 0.5ul DpnI |
Single Reaction:
- Add 250ng of DNA to be digested, and adjust with dH20 for a total volume of 16ul.
- Add 2.5ul of NEBuffer 2.
- Add 0.5ul of BSA.
- Add 0.5ul of EcoRI.
- Add 0.5ul of PstI.
- There should be a total volume of 20ul. Mix well and spin down briefly.
- Incubate the restriction digest at 37C for 30min, and then 80C for 20min to heat kill the enzymes. We incubate in a thermal cycler with a heated lid
- Run a portion of the digest on a gel (8ul, 100ng), to check that both plasmid backbone and part length are accurate.
Our Experience
Ligation Lab
Materials
Procedure
- Add 2ul of digested plasmid backbone (25 ng)
- Add equimolar amount of EcoRI-HF SpeI digested fragment (< 3 ul)
- Add equimolar amount of XbaI PstI digested fragment (< 3 ul)
- Add 1 ul T4 DNA ligase buffer. Note: Do not use quick ligase
- Add 0.5 ul T4 DNA ligase
- Add water to 10 ul
- Ligate 16C/30 min, heat kill 80C/20 min
- Transform with 1-2 ul of product
Our Experience
Transformation Lab
Materials
- Resuspended DNA
- Competent Cells
- Ice
- 2ml tube
- 42ºC water bath
- SOC media
- Petri dishes with LB agar and ampicillin
- glass spreader
- 37ºC incubator
- 10pg/ul RFP Control
Procedure
- Add 50 µL of thawed competent cells into pre-chilled 2ml tube, and another 50µL into a 2ml tube, labelled for your control.
- Add 1 - 2 µL of the resuspended DNA to the 2ml tube. Pipet up and down a few times. Make sure to keep the competent cells on ice.
- Add 1 µL of the RFP Control to control transformation.
- Close tubes and incubate the cells on ice for 30 minutes.
- Heat shock the cells by immersion in a pre-heated water bath at 42ºC for 60 seconds.
- Incubate the cells on ice for 5 minutes.
- Add 200 μl of SOC media (make sure that the broth does not contain antibiotics and is not contaminated) to each transformation.
- Incubate the cells at 37ºC for 2 hours while the tubes are rotating or shaking.
- Label two petri dishes with LB agar and ampicillin with the part number, plasmid backbone, and antibiotic resistance. Plate 20 µl and 200 µl of the transformation onto the dishes, and spread. This helps ensure that you will be able to pick out a single colony.
- For the control, label two petri dishes with LB agar (AMP). Plate 20 µl and 200 µl of the transformation onto the dishes, and spread.
- Incubate the plates at 37ºC for 12-14 hours, making sure the agar side of the plate is up.
- Count the colonies on the 20 μl control plate and calculate competent cell efficiency.
Our Experience
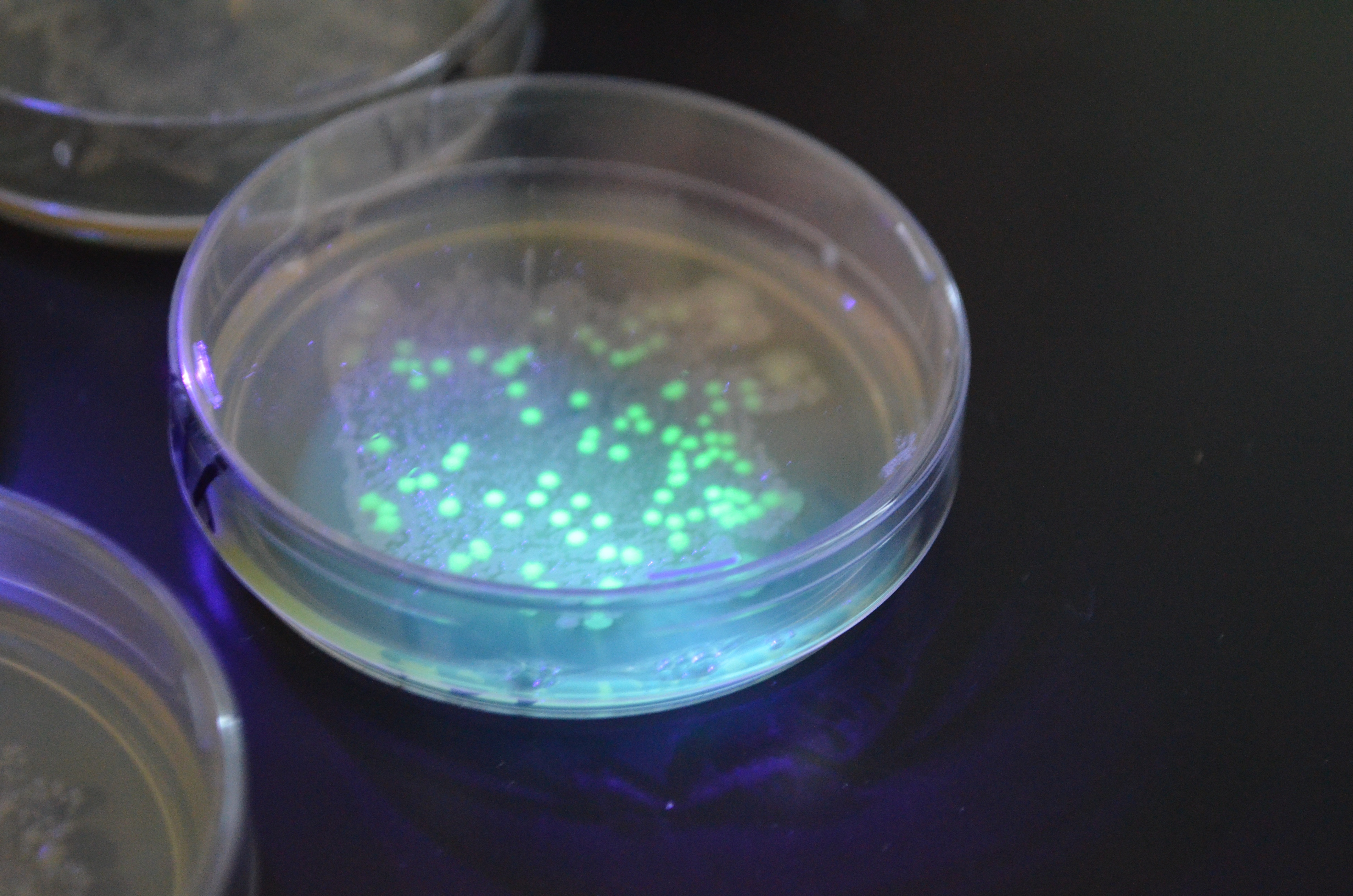
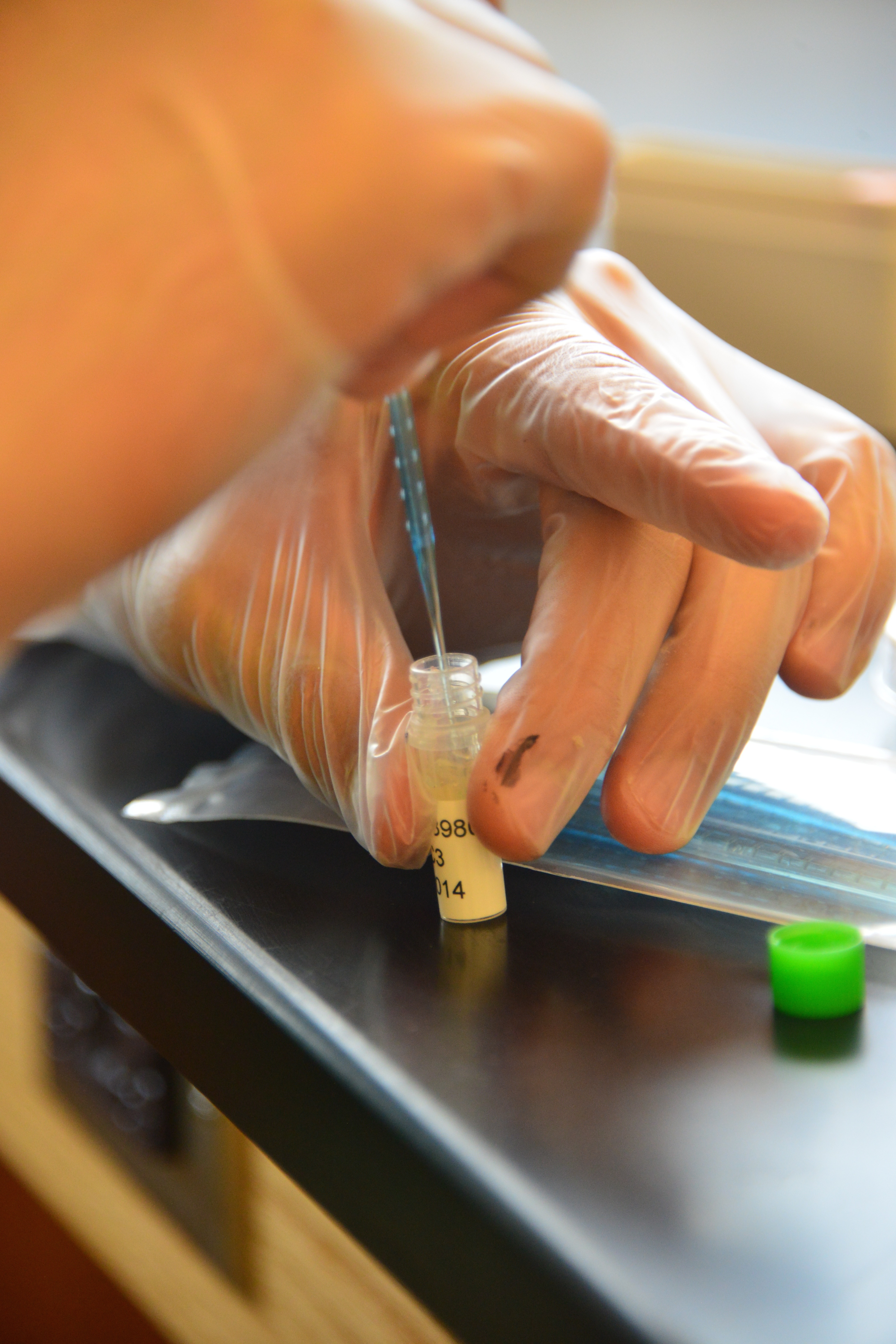
Lab Photobook
 "
"