Team:WalthamHS BioHawks/Project/Procedure
From 2014hs.igem.org
Procedure
Our lab procedures consisted of three activities; practice with the 3A assembly protocols, building our stain remover device, and testing the stain remover functionality.
Practicing with the 3A Assembly Kit
Before we began our project, we practiced using the 3A Assembly Kit: restriction digests, gel electrophoresis, ligation, and transformation.
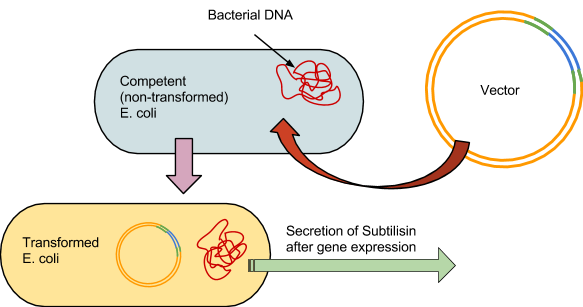
Building the Stain Remover
As with the 3A assembly practice, there were 4 fundamental steps to the process. The first was to use the restriction enzymes to digest the component parts, in this case BBa_J04500 and the synthesized subtilisin DNA. Our subtilisin gene and terminator were synthesized by BioBasics, and delivered in a plasmid with compatible BioBrick restriction sites. Second, a gel is run prior to ligation to verify the fragment sizes match the calculated predictions.The third was to ligate the two parts into a plasmid. And the fourth was to transform the E.coli with the plasmid. The engineered bacteria can then be incubated, allowing it to secrete subtilisin, which can then be extracted for potential stain removal.
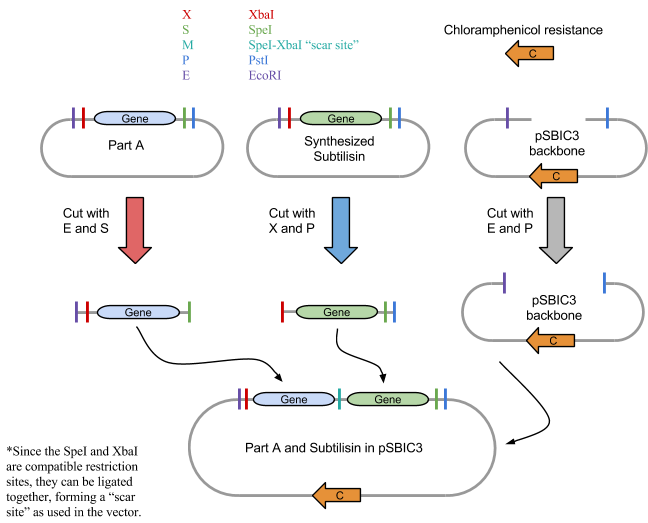
1: Restriction Digest
We first digested BBa_J04500, subtilisin, pSB1C3 and RFP control with their respective restriction enzymes. EcoRI and SpeI were added to BBa_J04500, XbaI and PstI were added to the synthesized subtilisin tube, and EcoRI and PstI to the plasmid pSBIC3 backbone tube. An RFP control tube received EcoRI and PstI, which would show that digestion worked properly. Each tube had approximately 50 ul of solution total and were incubated for 30 minutes at 37 degrees C and 20 minutes at 80 degrees C using a water bath.
2: Running the Gel
After performing the restriction digest for BBa_J04500, the subtilisin gene, the plasmid, and the RFP control, a gel was run to identify the fragment sizes and verify the digest occurred properly. The gel lanes were loaded using the DNA and loading dye, totaling 5 ul. The gel ran for 6 mins under 275v. If the digestion worked correctly, and the gel supplied evidence to support it, then the transformation could continue. However, after running the gel electrophoresis of all digested sequences, no visible results were found. We then repeated the digest and gel and still we were unsuccessful. View Our Results
Because the DNA ladder was present and its fragments were correctly distributed, the lack of data from the samples infered a digestion-based error of some kind. Without the vector, the experimental E. coli could not be grown, preventing the biological production of subtilisin. Due to the ultimately insufficient resources of DNA, we were unable to continue our transformation.
We hypothesized that the restriction digest error may have been caused by nonfunctional restriction enzymes. To test this, we digested DNA from the DNA distribution kit with restriction enzymes. Four samples of DNA was incubated each with one type of restiction enzyme. We ran the results on a gel, and the DNA did not appear to be digested. We concluded that the restriction enzymes were nonfunctional. View Our Results
3: Ligation into Plasmid
If digestion worked properly, the parts would be combined in solution and ligated. Although some variations of ligation would be arranged incorrectly, there should be sufficient correct vectors that could be inserted into E. coli.
4: Prokaryotic Transformation
The competent E. coli would be incubated on ice for half an hour. The vectors would then be forced in by heating the cells in a water bath at 47 degrees Celsius for 1 minute, then put back on ice for 5 minutes.
The bacteria are then grown at 37 degrees Celsius in an incubator for 24 hours on agar plates. Following initial incubation, they would then undergo another 24 hour growing period at room temperature in a controlled environment.
An antibiotic would be applied, and all bacteria not properly transformed would not grow, leaving only the subtilisin-producing colonies. The colonies are then placed in 5ml tubes, where the subtilisin will be secreted into solution and then extracted from the cells.
Testing the Stain Remover
Even though our transformation of E. coli failed, we still wanted to simulate how our project idea would have worked if the experiment was successful. Since our goal was to transform the subtilisin gene into E. coli to produce enzymes that digest proteins, we re-hydrated subtilisin enzyme powder (Sigma Aldrich P5380-25 mg) and tested its effectiveness on milk. We chose skim milk because it had a high concentration of proteins.
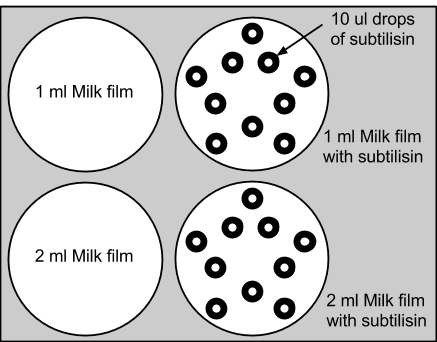
1: Initial Subtilisin Control Test
We added 1ml and 2ml of skim milk to agar plates, and placed 10 drops of 10ul of subtilisin enzyme onto each experimental dish. Subtilisin's enzymatic decomposition was successfully observed. The control plates with only milk were relatively unchanged. This experiment supported the functionality of subtilisin in the decomposition of the proteins. View Our Results
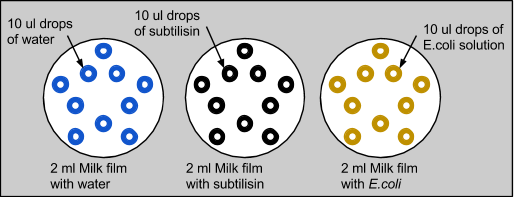
2: Complete Biological Control Test
We then wanted to confirm that subtilisin had protein-degrading capabilities, and not E. coli. We also wanted to eliminate liquid dispersion as a source of error. So, we repeated the experiment with three plates, each with 2ml of skim milk applied on top. The first control plate had 10 drops of 10ul of water, the second with 10 drops of 10ul of subtilisin, and the third with 10 drops of 10ul of E. coli, which was grown in LB overnight. Similar to the previous experiment with only milk, the control with water was unchanged and the water diffused into the milk without modifying it. When E. coli was applied to the milk, there were no significant changes. Similar to the last experiment, milk treated with subtilisin was degraded. This proved that subtilisin was solely responsible for protein degradation.
 "
"