Team:WalthamHS BioHawks/Project/Design
From 2014hs.igem.org
| Line 43: | Line 43: | ||
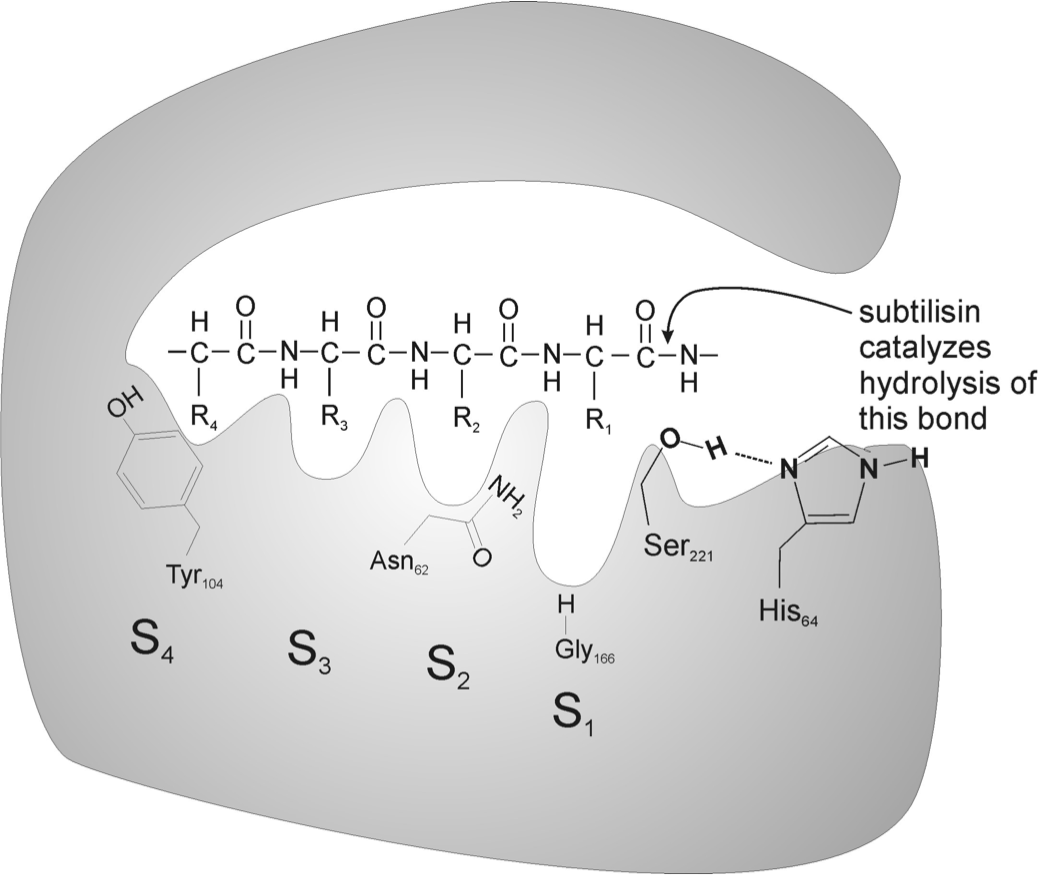
Subtilisin is a protein-degrading enzyme. The peptide bonds are degraded with serine residue at the active site. The enzyme works in a charge-relay network, using Asp-32, His-64, and Ser-221. The three amino acids form a group in the 3D shape of the protein as is indicated in Figure 1. Ser-221 is able to cleave the peptide bond of proteins with its partially negative oxygen atom as is shown in Figure 2. In commercial use, a genetically engineered form of subtilisin is used, as harsh chemical detergents and high temperatures easily inactivate the wild type. It usually consists of 269 to 275 amino acids and its active from pH 6 to 11, with its major activity from pH 9 to 11. Subtilisin comes from the Bacillus genus and is easily found in the environment (in soil bacteria) and is readily biodegradable. | Subtilisin is a protein-degrading enzyme. The peptide bonds are degraded with serine residue at the active site. The enzyme works in a charge-relay network, using Asp-32, His-64, and Ser-221. The three amino acids form a group in the 3D shape of the protein as is indicated in Figure 1. Ser-221 is able to cleave the peptide bond of proteins with its partially negative oxygen atom as is shown in Figure 2. In commercial use, a genetically engineered form of subtilisin is used, as harsh chemical detergents and high temperatures easily inactivate the wild type. It usually consists of 269 to 275 amino acids and its active from pH 6 to 11, with its major activity from pH 9 to 11. Subtilisin comes from the Bacillus genus and is easily found in the environment (in soil bacteria) and is readily biodegradable. | ||
| - | [[File:Protein1.png|left|500px]] [[File:Protein2.png|right]] | + | [[File:Protein1.png|left|500px]] [[File:Protein2.png|right|500px]] |
Revision as of 15:30, 19 June 2014
| Home | History of Waltham -Outreach |
Team | Project -Design |
Sponsors | Our Poster |
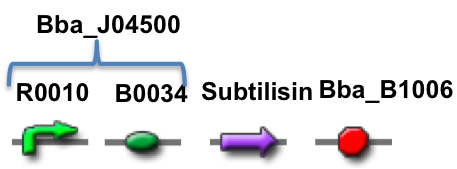
Our stain remover device is built of four main components; a promoter, ribosome binding site (RBS), the subtilisin enzyme and a terminator. The promoter and RBS are taken from the distribution kit under part Bba_J04500. We had the subtilisin enzyme and terminator synthesized by the company BioBasic at reasonable cost. In addition, they synthesized the Biobrick prefix and suffix with the part to make it 3A compatible.
Basic Descriptions:
The promoter and RBS are together in BioBrick Part BBa_J04500, which is the same part used in the practice 3A assembly kit. BB-prefix is the BioBrick sequence with the appropriate restriction enzyme sites to conform to the 3A assembly standard. OmpA is a short signal peptide that coaxes E.coli to secrete the downstream protein out of the cell. Pro-sequence is a protein that helps guide the folding of the subtilisin enzyme and is part of the natural subtilisin gene. Subtilisin is a serine protease enzyme that degrade other proteins. BB-suffix is the BiobBick sequence with the appropriate restriction enzyme sites to conform to the 3A Assembly standard. The terminator is BBa_B1006.
Basic Descriptions:
The promoter and RBS are together in BioBrick Part BBa_J04500, which is the same part used in the practice 3A assembly kit. Subtilisin is a serine protease enzyme that can degrade other proteins. The terminator is BBa_B1006.
Parts Explained:
The promoter is taken from the lac operon, which is inducible. A lac operon is a series of genes that code for proteins that digest lactose. However we have replaced those genes with genes that code for subtilisin. LacI codes for repressors that halt transcription, preventing RNA polymerase from binding. It can be regulated because it is CAP sensitive. It has two binding sites, the first allows the CAP protein to bind, while the second binds to the LacI protein. The IPTG is a molecular mimic of lactose. The gene can only be turned on only when IPTG is present, but when glucose is absent. RBS is the ribosome binding site, where the ribosomes bind together to begin translation. Our goal was to translate a protein subtilisin, a serine protein enzyme responsible for digesting protein-based molecules. Before the operon, there is a BB-prefix. After the termination sequence there is a BB-suffix, used to cease transcription of the subtilisin genes. The two BB sequences are restriction sites where the the gene can be cut to be incorporated into a plasmid. The promoter and RBS together are called Part A (BBa_J04500), totaling to 220 base pairs. Since the operon we are used was inducible, when is glucose, the accumulation of cAMP binds to CAP, which in turn allows transcription to occur. When LacI is absent, the repressors are not produced, so transcription is possible. If our goal is for subtilisin to be produced, then LacI and glucose should be absent, but IPTG should be present.
The specific subtilisin enzyme (Subtilisin BPN’, UniProt ID P00782 6 ) we used has been commercially marketed by the company Novozymes under the name Alcalase for use in detergents. Based on the work of Ghasemi (2012) and Yamabhai (2008) we knew that to get E. coli to excrete our enzyme a signal peptide would have to be added. After our advisor consulted with colleagues, the OmpA signal peptide was chosen. OmpA signal peptide is 21 amino acids long. It helps secrete recombinant enzymes out of the bacteria, where time is an important factor in how much is secreted. Induction time requires several hours, 4 in the experiment by Yamabhai. Normally, it is used to translocate and direct enzymes across the cell membrane of the bacteria. The type of bacteria should not be a major factor, but should also not be ignored. For some enzymes tested by Yamabhai, OmpA was equally effective in secretion as the native signal peptides, however this is not guaranteed for all enzymes. Due to the large variety of enzymes that OmpA can direct, it is advantageous to use it for non-native enzyme secretion in transformed E. coli bacteria.
Subtilisin is a protein-degrading enzyme. The peptide bonds are degraded with serine residue at the active site. The enzyme works in a charge-relay network, using Asp-32, His-64, and Ser-221. The three amino acids form a group in the 3D shape of the protein as is indicated in Figure 1. Ser-221 is able to cleave the peptide bond of proteins with its partially negative oxygen atom as is shown in Figure 2. In commercial use, a genetically engineered form of subtilisin is used, as harsh chemical detergents and high temperatures easily inactivate the wild type. It usually consists of 269 to 275 amino acids and its active from pH 6 to 11, with its major activity from pH 9 to 11. Subtilisin comes from the Bacillus genus and is easily found in the environment (in soil bacteria) and is readily biodegradable.
 "
"