Team:WalthamHS BioHawks/Project
From 2014hs.igem.org
Project
Throughout the fall and into the winter, our team created a list of many possible topics for our iGEM project. We narrowed our list down to five possible subjects: a pH sensor for tap water at our school, an environmentally friendly paper towel degrader, a stain remover for protein based stains, a pre-food taster that could detect any possible pathogens, and a more effective anti-allergy molecule. After a long discussion of the pros and cons of each topic, the team narrowed the debate to two topics: the pH sensor and the stain remover. There was much lobbying back and forth before the majority of people voted for the stain removal project. And so began our journey.
Our project was to create an environmentally friendly alternative to the harsh chemicals in modern detergents. More and more people every day are developing negative side effects from the continuous use of the damaging chemicals. Laundry detergents can cause rashes, red and blistered skin, sun sensitivity, sneezing, and itchy or watery eyes. Our goal was to create a powerful stain remover that was safe to use for all people.
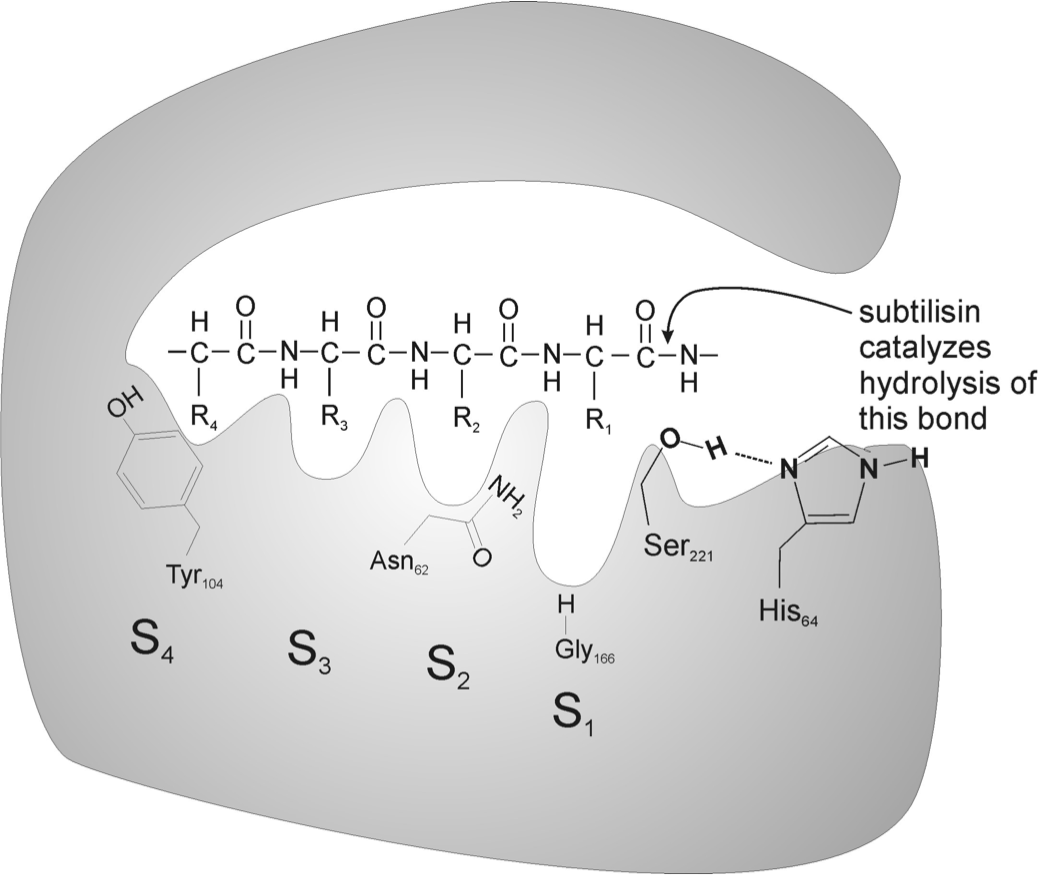
Each team member was given a part of the problem to research. Some looked at ingredients in common detergents, others researched natural stain removing remedies. In the end, we discovered a protease enzyme called subtilisin, a broad range protein degrader from the genus of bacteria Bacillus. Subtilisin is an enzyme found in many common laundry detergents and dish washers, but also found in nature.[ref 1] It can be obtained in certain soil bacteria, which creates it in large amounts.[ref 2] Subtilisin is made up of approximately 269-275 amino acids, with a typical globular shape. The active site of subtilisin involves Asp-32, His-64, and most importantly Ser-221. These three amino acids converge in a 3D shape to create the active site (see Figure 1.) The Ser-221 cleaves peptide bonds with its partially negative oxygen.[ref 3] (see Figure 2)
Our task was to transform the subtilisin enzyme into E. coli to create a bacteria that could rapidly produce many of the needed enzymes. E. coli has a reproduction rate of 15-20 minutes, which makes it able to quickly create daughter cells.[ref 4] This allows for a flexible bacterial host that is able to quickly test new enzyme designs.
Subtilisin also has the capability to break down blood clots.[ref 5] Blood clots occurs when blood platelets mature and combine with a protein, fibrin, to form a liquid-like gel. The blood clots occur naturally in the circulatory system to prevent blood loss in damaged blood vessels and open cuts. However, the coagulation of blood can be irregular, which can be serious if developed near or in the lungs, spine, or brain. Irregular clotting can be caused by things heart problems, stroke, obesity, and smoking. Subtilisin enzymes can be used to break down potentially harmful clots. The protease nature of the enzyme can be used to hydrolyze the fibrin (an insoluble protein). Subtilisin blocks the binding site of the fibrin to prevent more clotting, as well as breaking down the peptide bonds that hold the protein together.
References:
1. Karl-Heinz Maurer, “Detergent Proteases”, Current Opinion in Biotechnology 2004, 15:330-334
2. F. Niehaus et al “Enzymes for the laundry industries: tapping the vast metagenomic pool of alkaline proteases”, Microbial biotechnology 2011, 4(6), 767-776
3. http://en.wikipedia.org/wiki/Subtilisin
4. Graznya E. Sroga et al “A Strategy for in vivo screening of Subtilisin E reaction specificity in E. coli periplasm” Biotechnology and Bioengineering, VOL 78, NO 7, June 30 2002
5. Younes Ghasemi et al “Cloning of a Fibrinolytic Enzyme (subtilisin) gene from Bacillus Subtilis in Escherichia coli”, Molecular Biotechnology, 2012 52:1-7
6. http://www.uniprot.org/uniprot/P00782
7. Montarop Yamabhai “Secretion of recombinant Bacillus hydrolytic enzymes using Eschericia coli expression systems” Journal of Biotechnology 133(2008) 50-57
Figure 1. Crystal structure of subtilisin showing catalytic triad, image from http://www.cryst.bbk.ac.uk/PPS95/course/7_tertiary/glob_enz.html
Figure 2. Hydrolytic action of subtilisin, image from http://lib.convdocs.org/docs/index-130501.html?page=5
 "
"