Team:Jefferson VA SciCOS/Project
From 2014hs.igem.org
| Line 64: | Line 64: | ||
---- | ---- | ||
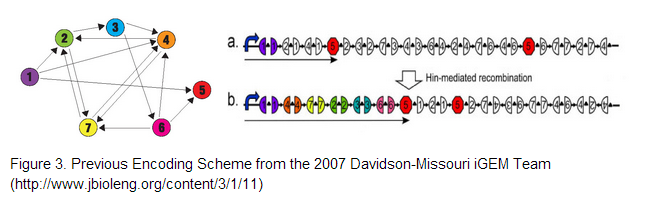
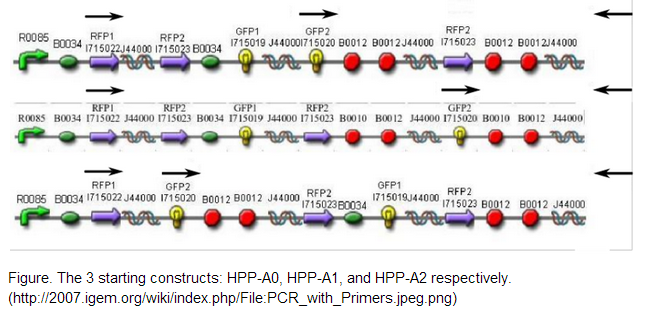
The 2007 Davidson/Western Missouri iGEM team constructed 3 initial pathways to demonstrate that the Hamiltonian Path Problem could be solved in vivo. <br /> <br /> | The 2007 Davidson/Western Missouri iGEM team constructed 3 initial pathways to demonstrate that the Hamiltonian Path Problem could be solved in vivo. <br /> <br /> | ||
| - | [[File:Encoding2.png]] | + | [[File:Encoding2.png]] <br /> <br /> |
We decided to use one of the starting constructs as a scaffold from which we could build our own composite pathway because each of the constructs contains three nodes: two split genes and a double terminator. Also, in the interest of time and resources, we were not concerned with constructing several pathways of our own as the Davidson/Western Missouri iGEM team had done. We ended up using HPP-A2 because it showed evidence of no fluorescence without Hin, which was the expected result as recorded by the Davidson/Western Missouri iGEM team. To HPP-A2, we added an RBS of moderate strength, as determined by the ribosome binding calculator and published values in literature. Compared to B0034, the RBS used in HPP-A2, B0032 is reported to exhibit 32% binding efficiency. Downstream of the RBS, we added another node: a BFP split-gene part. Thus, the order of our final part, from 5’ to 3’, was HPP-A2 + RBS + BFP1 + hixC + BFP2. We wanted to transform E.coli containing this composite pathway with Hin recombinase in order to shuffle the gene halves randomly and with varied efficiency, resulting in a solution to the TSP. | We decided to use one of the starting constructs as a scaffold from which we could build our own composite pathway because each of the constructs contains three nodes: two split genes and a double terminator. Also, in the interest of time and resources, we were not concerned with constructing several pathways of our own as the Davidson/Western Missouri iGEM team had done. We ended up using HPP-A2 because it showed evidence of no fluorescence without Hin, which was the expected result as recorded by the Davidson/Western Missouri iGEM team. To HPP-A2, we added an RBS of moderate strength, as determined by the ribosome binding calculator and published values in literature. Compared to B0034, the RBS used in HPP-A2, B0032 is reported to exhibit 32% binding efficiency. Downstream of the RBS, we added another node: a BFP split-gene part. Thus, the order of our final part, from 5’ to 3’, was HPP-A2 + RBS + BFP1 + hixC + BFP2. We wanted to transform E.coli containing this composite pathway with Hin recombinase in order to shuffle the gene halves randomly and with varied efficiency, resulting in a solution to the TSP. | ||
|} | |} | ||
Revision as of 23:53, 20 June 2014
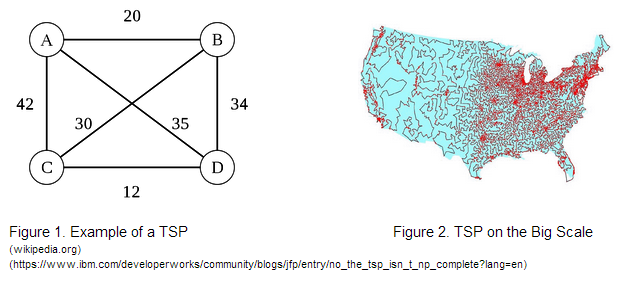
Solving a 4-Node Traveling Salesman Problem Using the Hin/hixC Recombinant SystemOur project’s goal was to assess the accuracy, scalability, and feasibility of a novel bacterial computer paradigm for solving a 4-node Traveling Salesman Problem. |
Bacterial Computing: The Future of the IT Industry All in an Agar PlateWhere would humanity be without the advent and rise of the computer? The 21st century has marked humanity’s entrance into a new Digital Age; now, more than ever, does the global marketplace and humanity itself depend on a thriving information technology sector. From the pocket-sized computing devices that we use to manage our day-to-day tasks to the massive supercomputers that mirror human intelligence, from the databases that store millions of patient’s records to the Big Data servers that analyze the vast number of financial transactions that take place every second, computing has evolved into a ubiquitous science that is both humanity’s present as well as its future. For years, the physical limitations that deterred the development of faster, more efficient computing devices were easily maneuverable, allowing computing to present itself as the seemingly perpetual gale force that propelled the sails of our global economy.
With these guiding questions in mind, our group has proposed a project that assesses the accuracy, scalability, and feasibility of a bacterial computer paradigm for solving different initial configurations of a 4-node Traveling Salesman Problem. Our work proposes a novel way of encoding the Traveling Salesman Problem using variable Ribosome Binding Site strengths and DNA constructs that can be solved by the Hin recombinase/HixC site system. |
Taking a Previous Encoding Scheme a Step FurtherThe 2007 Davidson-Missouri iGEM team proposed three encoding schemes given by Figure 3, where cities correspond to given genes and adjacent gene halves separated by hixC sites represent edges on the graph. To encode our 4-node problem we decided on using genes encoding for (1). blue fluorescent protein , (2) green fluorescent protein , (3) red fluorescent protein and a (4) terminator.
|
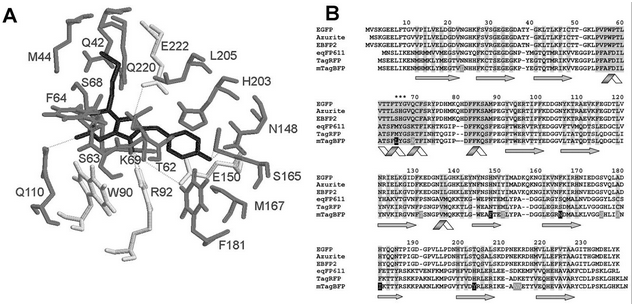
Engineering Our BFP ConstructIn order to carry out the encoding scheme we developed above, it was necessary to develop our own BioBrick to represent the two gene halves encoding for BFP. It was important to make sure that the protein’s fluorescent activity would not be compromised once the hixC site was inserted, i. e. our insertion could not occur within the BFP’s chromophore sequence. To discover what the chromophore sequence was for BFP, we read the documentation for Part:BBa_K592100 (mTagBFP as shown in the figure) as described by Subach et al., 2005 (http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2585067/). Figure from Subach et al., 2005. Letters specifying chromophore sequence are in black After having found the sequence for BFP and taking due note of which region encoded the chromophore, we then performed an analysis through the online gene splitting tool provided by the 2007 Davidson-Missouri Team (http://gcat.davidson.edu/iGEM07/genesplitter.html) in order to determine the correct primer to order. The following is a selection of the output from our analysis: |
The Hin Recombinase/hixC Site SystemThe Hin Recombinase/hixC Site System is what breathes life into the encoding scheme we designed above. The system designed by a 2006 iGEM team is derived from Salmonella, where Hin regulates how flagellin genes are expressed. In our project, the system was used to constitutively shuffle the gene halves until a solution is arrived at. |
 "
"