Team:CSWProteens/project
From 2014hs.igem.org
| (31 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | <html xmlns="http://www.w3.org/1999/xhtml"> | + | <html> |
| + | <style> | ||
| + | #contentSub, #footer-box, #catlinks, #search-controls, #p-logo, .printfooter, .firstHeading,.visualClear {display: none;} /*-- hides default wiki settings --*/ | ||
| + | </style><html xmlns="http://www.w3.org/1999/xhtml"> | ||
<head> | <head> | ||
<meta content="en-ca" http-equiv="Content-Language" /> | <meta content="en-ca" http-equiv="Content-Language" /> | ||
<meta content="text/html; charset=utf-8" http-equiv="Content-Type" /> | <meta content="text/html; charset=utf-8" http-equiv="Content-Type" /> | ||
| - | + | <script src="http://code.jquery.com/jquery-1.9.1.min.js"></script> | |
| + | <script src="https://2014hs.igem.org/Team:CSWProteens/jquery.slides.min?action=raw&ctype=text/javascript"></script> | ||
<style type="text/css"> | <style type="text/css"> | ||
| + | .auto-style1 { | ||
| + | font-family: Arial; | ||
| + | font-size: 12pt; | ||
| + | margin-top: 0px; | ||
| + | margin-bottom: 0px; | ||
| + | margin-right: 0px; | ||
| + | margin-left: 0px; | ||
| + | color: #000000 | ||
| + | } | ||
| + | |||
.auto-style2 { | .auto-style2 { | ||
| - | + | text-align: center; | |
} | } | ||
| + | |||
.auto-style3 { | .auto-style3 { | ||
| - | font-size: | + | text-align: center; |
| - | + | font-family: Arial; | |
| + | font-size: 50pt; | ||
| + | margin-top: 26px; | ||
| + | margin-bottom: 0px; | ||
| + | color: #058C1D; | ||
} | } | ||
| + | |||
.auto-style4 { | .auto-style4 { | ||
| - | color: #058C1D; | + | text-align: center; |
| + | font-family: Arial; | ||
| + | font-size: 19pt; | ||
| + | margin-top: 0px; | ||
| + | margin-bottom: 0px; | ||
| + | color: #058C1D; | ||
} | } | ||
| - | + | ||
| - | + | .auto-style5 { | |
| + | margin-top: 0px | ||
} | } | ||
| - | + | ||
| - | color: # | + | .auto-style6 { |
| + | font-family: Arial; | ||
| + | font-size: 20pt; | ||
| + | margin-top: 10px; | ||
| + | margin-bottom: 10px; | ||
| + | color: #000000; | ||
| + | |||
| + | .auto-style7 { | ||
| + | font-family: Arial; | ||
| + | font-size: 20pt; | ||
| + | margin-top: 5px; | ||
| + | margin-bottom: 5px; | ||
| + | margin-right: 0px; | ||
| + | margin-left: 0px; | ||
} | } | ||
| - | + | ||
| - | + | #content{ | |
| + | background-image:url(http://upload.wikimedia.org/wikipedia/commons/a/ac/White_flag_of_surrender.svg); | ||
} | } | ||
| - | . | + | |
| - | + | ||
| + | </style> | ||
| + | |||
| + | <style type="text/css"> | ||
| + | @import url(http://fonts.googleapis.com/css?family=Arial); | ||
| + | ul, li { | ||
| + | z-index:999; | ||
| + | margin: 0; | ||
| + | padding: 0; | ||
} | } | ||
| - | + | ||
| - | + | #nav li > a { | |
| - | + | transition: all 0.2s ease-in-out; | |
| - | + | ||
| - | + | ||
| - | + | ||
} | } | ||
| - | + | ||
| - | + | #nav { | |
| - | + | -webkit-box-shadow: 0px 0px 5px 3px rgba(0, 0, 0, 0.75); | |
| - | + | -moz-box-shadow: 0px 0px 5px 3px rgba(0, 0, 0, 0.75); | |
| - | + | box-shadow: 0px 0px 5px 3px rgba(0, 0, 75, 0.75); | |
| - | + | font-family: 'Dosis', sans-serif; | |
| - | + | font-size: 24px; | |
| + | text-align:center; | ||
| + | background-color: #ffffff; | ||
} | } | ||
| - | + | #nav a { | |
| - | + | color: #000000; | |
| - | + | text-decoration: none; | |
} | } | ||
| - | + | #nav li { | |
| - | + | display: inline; | |
| - | + | position: relative; | |
| - | + | ||
| - | + | ||
} | } | ||
| - | + | #nav li ul { | |
| - | + | position: absolute; | |
| - | + | left: -9999px; | |
} | } | ||
| - | + | #nav li a { | |
| - | + | display: inline-block; | |
| - | + | padding: 20px; | |
} | } | ||
| - | + | #nav li:hover a { | |
| - | + | background-color: #000000; | |
| - | + | color: #ffffff; | |
| - | + | ||
| - | + | ||
| - | + | ||
} | } | ||
| - | + | #nav li:hover ul { | |
| - | + | left: 0; | |
| - | + | text-align: left; | |
| - | + | width: 200px; | |
| - | + | box-shadow: 0px 1px 1px #20d681; | |
| - | + | ||
} | } | ||
| - | + | #nav li:hover ul a { | |
| - | + | display: block; | |
| + | padding: 15px 30px 15px 30px; | ||
| + | border-bottom: 1px solid #70a667; | ||
} | } | ||
| - | # | + | #nav li:hover ul a:hover { |
| - | background- | + | background-color: #70a667; |
| + | color: #40004b; | ||
} | } | ||
</style> | </style> | ||
| Line 86: | Line 130: | ||
<meta content="revealTrans(Duration=3.0,Transition=2)" http-equiv="Site-Enter" /> | <meta content="revealTrans(Duration=3.0,Transition=2)" http-equiv="Site-Enter" /> | ||
</head> | </head> | ||
| + | <center><a href="https://2014hs.igem.org/Team:CSWProteens/project"><img src="https://static.igem.org/mediawiki/2014hs/7/7e/CSW_Banner.png" width="600"> | ||
| + | </a> | ||
| - | < | + | <a href="https://2014hs.igem.org/Main_Page"><img alt="" class="auto-style5" height="200" src="https://static.igem.org/mediawiki/2014hs/2/26/Igem_grpyon.png" width="200" /></a> </p></center> |
| - | < | + | <body style=" color: #FFFFFF;background-color: #000000; |
| - | + | background-image: url('http://upload.wikimedia.org/wikipedia/commons/a/ac/White_flag_of_surrender.svg');"> | |
| - | </ | + | <ul id="nav"> |
| - | </ | + | <li><a href="https://2014hs.igem.org/Team:CSWProteens"><b>Home</b></a></li> |
| - | + | <li> | |
| - | </ | + | <a href="https://2014hs.igem.org/Team:CSWProteens/team"><b>Team</b></a> |
| - | < | + | <ul> |
| - | </ | + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/team">Members</a></li> |
| + | <li><a href="http://www.csw.org/">The Cambridge School of Weston</a></li> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/advisors">Advisors</a></li> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/sponsors">Sponsors</a></li> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/Attributions">Attributions</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="https://2014hs.igem.org/Team:CSWProteens/project"><b>Project</b></a> | ||
| + | <ul> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/project/frostprotection">Frost Protection</a></li> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/project">Project Description</a></li> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/project/Methods">Methods</a></li> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/project/results">Results</a></li> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/project/discussion">Discussion</a></li> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/project/safety">Safety</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="https://2014hs.igem.org/Team:CSWProteens/notebook"><b>Notebook</b></a> | ||
| + | <ul> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/notebook/">Lab Journal</a></li> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/notebook/protocols">Protocols</a></li> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/notebook/parts">Parts</a></li> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/notebook/references">References</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="https://2014hs.igem.org/Team:CSWProteens/humanpractices"><b>Outreach</b></a> | ||
| + | <ul> | ||
| + | |||
| + | </ul> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="https://2014hs.igem.org/Team:CSWProteens/contact"><b>Contact Us</b></a> | ||
| + | <ul> | ||
| + | <li><a href="http://csw-igem.tumblr.com">Tumblr</a></li> | ||
| + | <li><a href="http://www.facebook.com/cswigem">Facebook</a></li> | ||
| + | <li><a href="mailto:hgoldsweig@csw.org?subject=iGEM">Email</a></li> | ||
| + | </ul> | ||
| + | </ul> | ||
| - | <center><img alt="" class="auto-style11" height=" | + | <center><b><p class="auto-style6" style="height: 30px">P R O J E C T D E S C R I P T I O N</b></p></center> |
| - | <p> | + | <center><img alt="" class="auto-style11" height="500" src="https://static.igem.org/mediawiki/2014hs/c/cd/Plantifreeze.gif" width="559" /></center><p> |
| - | < | + | <p><p class="auto-style1"> |
| - | + | Annually, about 5% to 15% of agricultural produce is lost due to frost. The formation of ice damages plants by rupturing cells and also through dehydration as water molecules are drawn out of the tissue. Current solutions to this problem - such as using heat or covering crops with protective material - are cumbersome, costly, and not fully preventative. Synthetic antifreeze chemicals have not been proven to work. Even if they did, they would be need to be applied repeatedly, at great cost, and may also leave residues in the environment. | |
| - | The CSW | + | <p><p class="auto-style1"> |
| - | <b> | + | The project undertaken by the Cambridge School of Weston (CSW) 2014 iGEM team aims to design a synthetic biology solution to this problem by building upon the work of the 2009 Utah State team that developed a protein secretion mechanism and that of the 2011 Yale team which synthesized an “antifreeze” protein (RiAFP) isolated from a cold-tolerant beetle called Rhagium inquisitor. This is the most potent antifreeze protein so far studied. Our device (Plantifreeze) is designed to mitigate crop damage caused by ice crystal formation on the plant surface. Plantifreeze is a dual plasmid system. |
| - | + | <p><p class="auto-style1"><b> | |
| - | + | Transport of RiAFP by the bacterium:</b> <p><p class="auto-style1">We quickly realized that expression of RiAFP in the cytoplasm was only half the functionality needed. The protein had to be secreted outside the cell envelop of the bacterium, so the surface of the crop would be protected from damage by ice crystals. E.coli have two membranes: inner membrane and outer membrane. To transport RiAFP protein to outside of E.coli, the protein has to pass through the two membranes. However, we were puzzled by how we would get the individual bacterium to transport the protein out of the bacterial cell. We could lyze each bacterium but this seems not to be a very elegant solution. So we needed to find a transport system for RiAFP.<p> | |
| + | <p class="auto-style1"> | ||
| + | Our research into the transport of proteins in E coli was very interesting. E. coli normally does not secrete proteins extra-cellularly except for a few classes of proteins such as toxins and hemolysin (certain proteins and lipids that cause lysis of red blood cells by damaging their cell membrane). There are six pathways for secretion of recombinant proteins in E coli, numbered I through VI. While all of these pathways differ mechanistically, they each promote secretion while maintaining the integrity of the cell structure. Types I and II are the most common pathways for recombinant protein secretion. | ||
| + | <p><p class="auto-style1"> | ||
| + | <b>Utah State to the rescue.</b> <p><p class="auto-style1">Further review of past iGEM projects led us to the work of the Utah State University iGEM 2009 team. As their project, they developed improved production and harvesting methods for proteins and other products in multiple organisms using the standardized BioBrick system. In short, they developed a library of fusion-compatible BioBrick parts for targeting compounds for secretion. | ||
| + | <p><p class="auto-style1"> | ||
| + | From their work we learned that recombinant proteins can be targeted to type I and II secretory pathways through genetic fusion with signal peptide targeting sequences. Type I secretion is a simple one step secretion system that can translocate proteins from the cytoplasm to the extracellular medium without protein interaction with the periplasm. | ||
| + | <p><p class="auto-style1"> | ||
| + | Asif Rahman from Utah State was extremely helpful following our contacting him by email. He explained, “It looks like a type I secretion system would be most useful for your group.” | ||
| + | <p><p class="auto-style1"><b> | ||
| + | The type 1 secretory system (T1SS). </b><p class="auto-style1">The alpha-hemolysin system is one of the best-studied type 1 secretion systems of E. coli. In T1SS the secretion occurs in a single step directly from the cytosol to the extracellular medium. The secretory machinery (translocator) of the alpha-hemolysin system consists of three proteins: C, an ATP binding cassette; HlyD, a membrane fusion protein; and TolC, an outer membrane protein. Proteins with C-terminally fused HlyA signal sequence can be recognized by the HlyB-HlyD-TolC translocator and targeted for secretion. So we fused the HlyA secretion tag to the C terminus end of the coding sequence for RiAFP. The expression of RiAFP fused with HlyA is controlled by one plasmid we call the expression plasmid. The translocator proteins (HlyB-HlyD-TolC) are expressed by a second compatible plasmid, pLG575. So the PlantiFreeze genetic circuit is a dual plasmid system consisting of the expression plasmid and the secretion plasmid, pLG575. These two plasmids had to be compatible (ie, different origins of replication and independent antibiotic selection). Plasmid pLG575 has p15A origin of replication and carries the chloramphenicol resistance marker. | ||
| + | <p><center><img src="https://static.igem.org/mediawiki/2014hs/f/f9/PLG575.png" width="350"><img src="https://static.igem.org/mediawiki/2014hs/2/2e/Secretion.jpg" width="450"></center><p class="auto-style1"> | ||
| + | <b> | ||
| + | Design of the expression plasmid. </b><p><p class="auto-style1">We decided to engineer a single DNA fragment to serve as the expression genetic circuit in the PlantiFreeze Device. The circuit comprises the following components: | ||
| + | <p><p class="auto-style1"> | ||
| + | BioBrick Standard Protein Coding Region Prefix /T7 Promoter /RBS /RiAFP /HlyA /Double terminator /BioBrick Standard Protein Coding Region Suffix | ||
| + | <p><p class="auto-style1"> | ||
| + | <b>Promoter. </b><p><p class="auto-style1">Our device uses a T7 system, as did the Yale team, because T7 RNA polymerase is an incredibly fast and powerful enzyme transcribing rapidly and profusely for as long as the T7 RNA polymerase is present. It synthesizes RNA at a rate several times that of E. coli RNA polymerase and it terminates transcription less frequently. | ||
| + | <p><p class="auto-style1"> | ||
| + | The T7 promoter is a BioBrick part BBa_1712074 | ||
| + | <p><p class="auto-style1"><b> | ||
| + | C-terminal His-tag. </b><p><p class="auto-style1">In our protein expression construct, there is an amino acid motif that consists of six histidine (His) residues fused to the C-terminus of RiAFP. Histidine tags are widely used because they are small and rarely interfere with the function, activity, or structure of target proteins. The polyhistidine-tag is to be used to detect the secreted protein via anti-polyhistidine-tag antibodies or alternatively by in-gel staining (SDS-PAGE) with fluorescent probes bearing metal ions. | ||
| + | <p><p class="auto-style1"><b> | ||
| + | HlyA-signal peptide. </b><p><p class="auto-style1">The HlyA is a signal peptide found in the C-terminal signal sequence of alpha-hemolysin (HlyA). It is used to target RiAFP for secretion via the Type I secretion pathway of gram-negative bacteria. Fusion of the HlyA signal peptide to RiAFP results in transport of the protein from the cytoplasm to the extracellular medium in a single step. | ||
| + | <p><p class="auto-style1"> | ||
| + | The HlyA signal peptide is BioBrick pat BBa_K208006 | ||
| + | <p><p class="auto-style1"><b> | ||
| + | Terminator. </b><p><p class="auto-style1">There are several E. coli transcriptional terminators available via the Registry. The most commonly used type of terminator is a forward terminator. When placed downstream of a genetic part that is transcribed, a forward transcriptional terminator will cause transcription to abort. We use part BBa_B0015, a double terminator (B0010-B0012). BBa_B0015. This terminator has an Average Forward Termination Efficiency of 98.4%.<p><center> | ||
| + | <img alt ="" class="auto-style11" height="300" src="https://static.igem.org/mediawiki/2014hs/0/01/Plasmid_1_Design.png" width="429"/></center> | ||
| + | <p><b> | ||
| + | <p class="auto-style1"> | ||
| + | Synthesis of the gBlock DNA fragment. </b><p><p class="auto-style1">The DNA sequences for the components could be found in the Registry. So it was not difficult to design the part. It was 864 base pairs in length. It is fast and simple to order a 864 base pair gBlock™ fragment from Integrated DNA Technologies (IDT) (double-stranded, sequence-verified genomic blocks that ship in only a few working days for affordable and easy gene construction). The first step in ordering a gBlock is automated review by screening tools and expert review by IDT scientists of entered sequences for characteristics that may interfere with synthesis. This review found that there were sequence repetitions longer than 8 base pairs that comprise a combined 91% of the sequences within a window of 70 base pairs beginning at base pair 235. This structure would interfere with synthesis. So the IDT gene product specialist optimized the sequence for expression in E coli and compatible with the gBlock synthesis process. Optimization theoretically just changes codon usage without changing the proteins coded. Below is the optimized sequence for our expression part: | ||
| + | <p><center><p class="auto-style1"> | ||
| + | gaattcgcgccctttactggtcatccgcttaacgattggaaacacctcctatctcttcttcctgcaca<br>cccgcgtgaaagaagttaaaaagatgtatagctgtcgtgcggttggtgtagatgggcgtgccgtca<br>ccgatattcaaggcacctgccacgcgaaggcaacgggggcgggtgcgatggctagcggtacctcgg<br>agccggggtcaacgtccaccgcgactgcgaccggccgtggcgctaccgcccggagcacaagtacaggccg<br>gggaacagccacgaccacggcaaccgggacagcgagcgccacgtctaacgcaatcggtcagggtacc<br>gcgactaccacagcgactgggtcagcaggcggtcgggccacaggttccgctacgacctccagttctgcatcc<br>cagccgacccagacccagactattacggggccgggctttcagaccgccaaatcttttgcacgcaacacgg<br>caactactaccgtgactgcgtctcatcaccatcatcaccatctggcgtatggttcacaaggtgacttaaacccgct<br>gatcaacgaaatttcaaagatcatttcagcggcgggttcctttgatgtgaaagaggaacgcaccgccgcc<br>agcttgctccagttatcggggaatgcgagcgattttagttatggccgtaactcgattacgctcaccacgtc<br>agcttaataatattaatcccaggcaagcaataaaactaagggttctgtggagcgccttggtctgtccttttacttact<br>gttcgttggtgagcgctcgttattggagtctcactggctgactttcggttgggcgtttttacgtctgtatac<br>aagctccggccgctgcag</center> | ||
| + | <p><center><a href="https://static.igem.org/mediawiki/2014hs/1/10/IGEM_Seqbuilder_Expression_plasmid_in_backbone.png"><img src="https://static.igem.org/mediawiki/2014hs/1/10/IGEM_Seqbuilder_Expression_plasmid_in_backbone.png" width="450"></a> <a href="https://static.igem.org/mediawiki/2014hs/6/61/Screen_shot_2014-06-13_at_11.04.23_AM.png"> <img src="https://static.igem.org/mediawiki/2014hs/6/61/Screen_shot_2014-06-13_at_11.04.23_AM.png" width="450"></a><br><p class="auto-style1"><b>Sequence outlined in SeqBuilder (click to enlarge)</b></center><p> | ||
| + | <p class="auto-style1">Importantly, the EcoRI and PstI sites on the 5’ and 3’ ends of this gene were maintained compatible with BioBrick standard 3A assembly. | ||
| + | <p><p class="auto-style1"> | ||
| + | This expression part ordered from IDT would be inserted into a linearized plasmid backbone with antibiotic resistance different from the plasmid controlling secretion (pLG575). We decided to use pSB1A3. This linearized plasmid is supplied by iGEM. We had exhausted our previous supply so ordered a new one. | ||
| + | <p><p class="auto-style1"> | ||
| + | The plan is to insert the expression part into pSB1A3 and do cotransformation of BL21 DE3 cells with the two plasmids. Then we would induce the cotransformants with IPTG to express and secrete RiAFP. Finally, using the Invision In-gel Staining Kit we would analyze the media and BL21 DE3 cell lyzate for RiAFP. This would mean we work up to the very last minute before the Jamboree but we are determined to successfully complete the project. | ||
| + | <p><p class="auto-style1"><b> | ||
| + | Why use BL21 DE3 cells for the expression platform? </b><p><p class="auto-style1">Expression can only be achieved in a bacterial strain carrying the gene for the T7 RNA polymerase (see paragraph on T7 promoter above). The most common cell strain to use with a T7 promoter system is BL21 (DE3) (competent cells are chemically competent cells used for high-level protein expression with T7 RNA polymerase-based expression systems). The BL21(DE3) contains the T7 RNA polymerase gene, under the control of the lacUV5 promoter, integrated into the chromosome. IPTG is used to induce the expression of recombinant proteins cloned into vectors downstream of a T7 RNA promoter and transformed into the BL21(DE3) cells | ||
| + | <p><p class="auto-style1"><b> | ||
| + | pLG575 plasmid. </b><p><p class="auto-style1">The pLG575 plasmid was generously sent to us by our advisor at Utah State, Asif Rahman. On Nanodrop, the plasmid showed 260 ug/ul. | ||
| - | < | + | </p> |
| + | <center> | ||
| + | <img src="https://static.igem.org/mediawiki/2014hs/c/ce/Project_Diagram.png" width="500"> | ||
Latest revision as of 02:18, 21 June 2014


P R O J E C T D E S C R I P T I O N

Annually, about 5% to 15% of agricultural produce is lost due to frost. The formation of ice damages plants by rupturing cells and also through dehydration as water molecules are drawn out of the tissue. Current solutions to this problem - such as using heat or covering crops with protective material - are cumbersome, costly, and not fully preventative. Synthetic antifreeze chemicals have not been proven to work. Even if they did, they would be need to be applied repeatedly, at great cost, and may also leave residues in the environment.
The project undertaken by the Cambridge School of Weston (CSW) 2014 iGEM team aims to design a synthetic biology solution to this problem by building upon the work of the 2009 Utah State team that developed a protein secretion mechanism and that of the 2011 Yale team which synthesized an “antifreeze” protein (RiAFP) isolated from a cold-tolerant beetle called Rhagium inquisitor. This is the most potent antifreeze protein so far studied. Our device (Plantifreeze) is designed to mitigate crop damage caused by ice crystal formation on the plant surface. Plantifreeze is a dual plasmid system.
Transport of RiAFP by the bacterium:
We quickly realized that expression of RiAFP in the cytoplasm was only half the functionality needed. The protein had to be secreted outside the cell envelop of the bacterium, so the surface of the crop would be protected from damage by ice crystals. E.coli have two membranes: inner membrane and outer membrane. To transport RiAFP protein to outside of E.coli, the protein has to pass through the two membranes. However, we were puzzled by how we would get the individual bacterium to transport the protein out of the bacterial cell. We could lyze each bacterium but this seems not to be a very elegant solution. So we needed to find a transport system for RiAFP.
Our research into the transport of proteins in E coli was very interesting. E. coli normally does not secrete proteins extra-cellularly except for a few classes of proteins such as toxins and hemolysin (certain proteins and lipids that cause lysis of red blood cells by damaging their cell membrane). There are six pathways for secretion of recombinant proteins in E coli, numbered I through VI. While all of these pathways differ mechanistically, they each promote secretion while maintaining the integrity of the cell structure. Types I and II are the most common pathways for recombinant protein secretion.
Utah State to the rescue.
Further review of past iGEM projects led us to the work of the Utah State University iGEM 2009 team. As their project, they developed improved production and harvesting methods for proteins and other products in multiple organisms using the standardized BioBrick system. In short, they developed a library of fusion-compatible BioBrick parts for targeting compounds for secretion.
From their work we learned that recombinant proteins can be targeted to type I and II secretory pathways through genetic fusion with signal peptide targeting sequences. Type I secretion is a simple one step secretion system that can translocate proteins from the cytoplasm to the extracellular medium without protein interaction with the periplasm.
Asif Rahman from Utah State was extremely helpful following our contacting him by email. He explained, “It looks like a type I secretion system would be most useful for your group.”
The type 1 secretory system (T1SS).
The alpha-hemolysin system is one of the best-studied type 1 secretion systems of E. coli. In T1SS the secretion occurs in a single step directly from the cytosol to the extracellular medium. The secretory machinery (translocator) of the alpha-hemolysin system consists of three proteins: C, an ATP binding cassette; HlyD, a membrane fusion protein; and TolC, an outer membrane protein. Proteins with C-terminally fused HlyA signal sequence can be recognized by the HlyB-HlyD-TolC translocator and targeted for secretion. So we fused the HlyA secretion tag to the C terminus end of the coding sequence for RiAFP. The expression of RiAFP fused with HlyA is controlled by one plasmid we call the expression plasmid. The translocator proteins (HlyB-HlyD-TolC) are expressed by a second compatible plasmid, pLG575. So the PlantiFreeze genetic circuit is a dual plasmid system consisting of the expression plasmid and the secretion plasmid, pLG575. These two plasmids had to be compatible (ie, different origins of replication and independent antibiotic selection). Plasmid pLG575 has p15A origin of replication and carries the chloramphenicol resistance marker.


Design of the expression plasmid.
We decided to engineer a single DNA fragment to serve as the expression genetic circuit in the PlantiFreeze Device. The circuit comprises the following components:
BioBrick Standard Protein Coding Region Prefix /T7 Promoter /RBS /RiAFP /HlyA /Double terminator /BioBrick Standard Protein Coding Region Suffix
Promoter.
Our device uses a T7 system, as did the Yale team, because T7 RNA polymerase is an incredibly fast and powerful enzyme transcribing rapidly and profusely for as long as the T7 RNA polymerase is present. It synthesizes RNA at a rate several times that of E. coli RNA polymerase and it terminates transcription less frequently.
The T7 promoter is a BioBrick part BBa_1712074
C-terminal His-tag.
In our protein expression construct, there is an amino acid motif that consists of six histidine (His) residues fused to the C-terminus of RiAFP. Histidine tags are widely used because they are small and rarely interfere with the function, activity, or structure of target proteins. The polyhistidine-tag is to be used to detect the secreted protein via anti-polyhistidine-tag antibodies or alternatively by in-gel staining (SDS-PAGE) with fluorescent probes bearing metal ions.
HlyA-signal peptide.
The HlyA is a signal peptide found in the C-terminal signal sequence of alpha-hemolysin (HlyA). It is used to target RiAFP for secretion via the Type I secretion pathway of gram-negative bacteria. Fusion of the HlyA signal peptide to RiAFP results in transport of the protein from the cytoplasm to the extracellular medium in a single step.
The HlyA signal peptide is BioBrick pat BBa_K208006
Terminator.
There are several E. coli transcriptional terminators available via the Registry. The most commonly used type of terminator is a forward terminator. When placed downstream of a genetic part that is transcribed, a forward transcriptional terminator will cause transcription to abort. We use part BBa_B0015, a double terminator (B0010-B0012). BBa_B0015. This terminator has an Average Forward Termination Efficiency of 98.4%.

Synthesis of the gBlock DNA fragment.
The DNA sequences for the components could be found in the Registry. So it was not difficult to design the part. It was 864 base pairs in length. It is fast and simple to order a 864 base pair gBlock™ fragment from Integrated DNA Technologies (IDT) (double-stranded, sequence-verified genomic blocks that ship in only a few working days for affordable and easy gene construction). The first step in ordering a gBlock is automated review by screening tools and expert review by IDT scientists of entered sequences for characteristics that may interfere with synthesis. This review found that there were sequence repetitions longer than 8 base pairs that comprise a combined 91% of the sequences within a window of 70 base pairs beginning at base pair 235. This structure would interfere with synthesis. So the IDT gene product specialist optimized the sequence for expression in E coli and compatible with the gBlock synthesis process. Optimization theoretically just changes codon usage without changing the proteins coded. Below is the optimized sequence for our expression part:
gaattcgcgccctttactggtcatccgcttaacgattggaaacacctcctatctcttcttcctgcaca
cccgcgtgaaagaagttaaaaagatgtatagctgtcgtgcggttggtgtagatgggcgtgccgtca
ccgatattcaaggcacctgccacgcgaaggcaacgggggcgggtgcgatggctagcggtacctcgg
agccggggtcaacgtccaccgcgactgcgaccggccgtggcgctaccgcccggagcacaagtacaggccg
gggaacagccacgaccacggcaaccgggacagcgagcgccacgtctaacgcaatcggtcagggtacc
gcgactaccacagcgactgggtcagcaggcggtcgggccacaggttccgctacgacctccagttctgcatcc
cagccgacccagacccagactattacggggccgggctttcagaccgccaaatcttttgcacgcaacacgg
caactactaccgtgactgcgtctcatcaccatcatcaccatctggcgtatggttcacaaggtgacttaaacccgct
gatcaacgaaatttcaaagatcatttcagcggcgggttcctttgatgtgaaagaggaacgcaccgccgcc
agcttgctccagttatcggggaatgcgagcgattttagttatggccgtaactcgattacgctcaccacgtc
agcttaataatattaatcccaggcaagcaataaaactaagggttctgtggagcgccttggtctgtccttttacttact
gttcgttggtgagcgctcgttattggagtctcactggctgactttcggttgggcgtttttacgtctgtatac
aagctccggccgctgcag
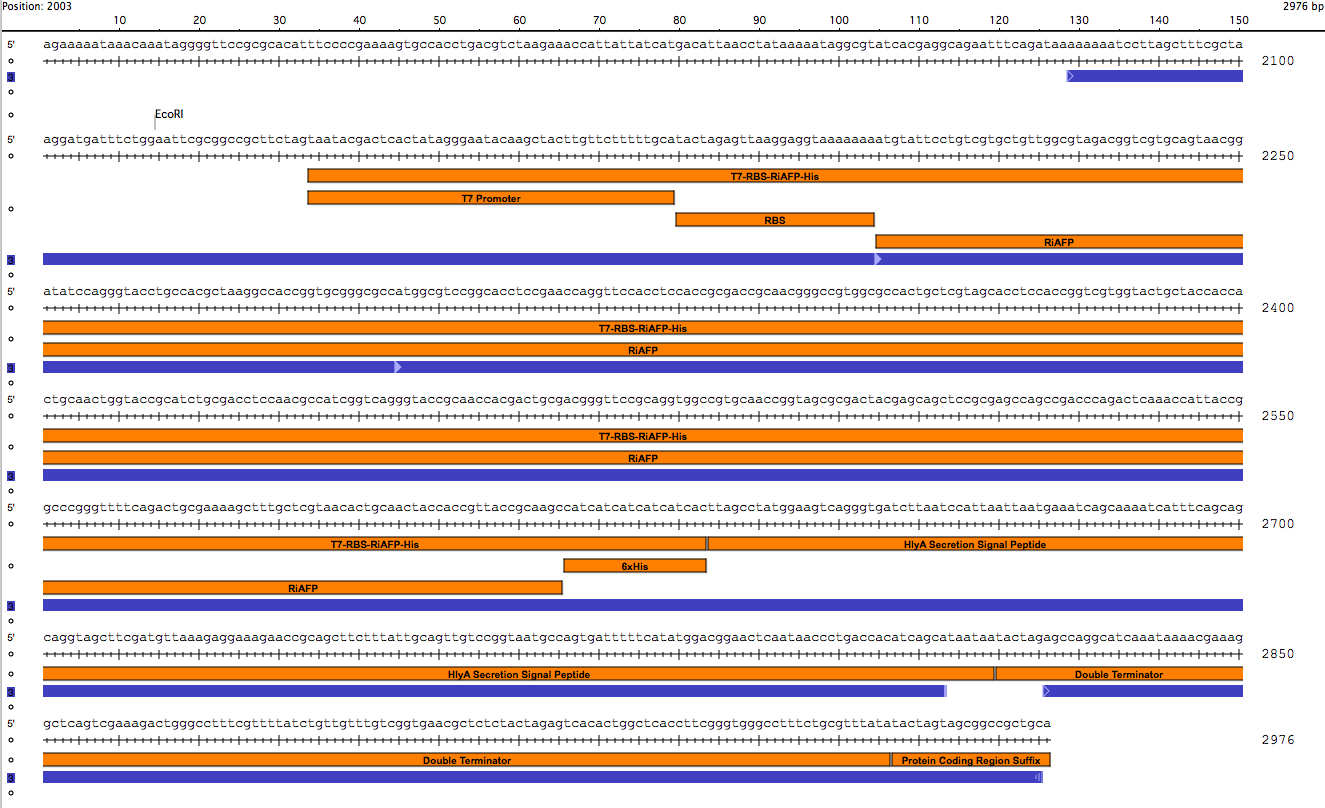
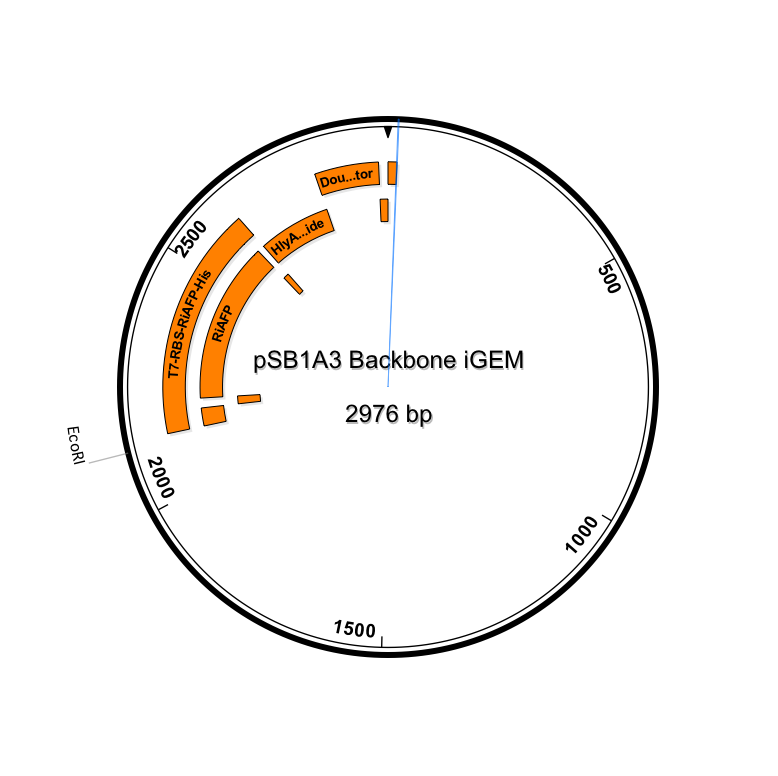
Sequence outlined in SeqBuilder (click to enlarge)
Importantly, the EcoRI and PstI sites on the 5’ and 3’ ends of this gene were maintained compatible with BioBrick standard 3A assembly.
This expression part ordered from IDT would be inserted into a linearized plasmid backbone with antibiotic resistance different from the plasmid controlling secretion (pLG575). We decided to use pSB1A3. This linearized plasmid is supplied by iGEM. We had exhausted our previous supply so ordered a new one.
The plan is to insert the expression part into pSB1A3 and do cotransformation of BL21 DE3 cells with the two plasmids. Then we would induce the cotransformants with IPTG to express and secrete RiAFP. Finally, using the Invision In-gel Staining Kit we would analyze the media and BL21 DE3 cell lyzate for RiAFP. This would mean we work up to the very last minute before the Jamboree but we are determined to successfully complete the project.
Why use BL21 DE3 cells for the expression platform?
Expression can only be achieved in a bacterial strain carrying the gene for the T7 RNA polymerase (see paragraph on T7 promoter above). The most common cell strain to use with a T7 promoter system is BL21 (DE3) (competent cells are chemically competent cells used for high-level protein expression with T7 RNA polymerase-based expression systems). The BL21(DE3) contains the T7 RNA polymerase gene, under the control of the lacUV5 promoter, integrated into the chromosome. IPTG is used to induce the expression of recombinant proteins cloned into vectors downstream of a T7 RNA promoter and transformed into the BL21(DE3) cells
pLG575 plasmid.
The pLG575 plasmid was generously sent to us by our advisor at Utah State, Asif Rahman. On Nanodrop, the plasmid showed 260 ug/ul.

 "
"