Team:CSWProteens/notebook
From 2014hs.igem.org
| (33 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | <html xmlns="http://www.w3.org/1999/xhtml"> | + | <html> |
| + | <style> | ||
| + | #contentSub, #footer-box, #catlinks, #search-controls, #p-logo, .printfooter, .firstHeading,.visualClear {display: none;} /*-- hides default wiki settings --*/ | ||
| + | </style><html xmlns="http://www.w3.org/1999/xhtml"> | ||
<head> | <head> | ||
<meta content="en-ca" http-equiv="Content-Language" /> | <meta content="en-ca" http-equiv="Content-Language" /> | ||
<meta content="text/html; charset=utf-8" http-equiv="Content-Type" /> | <meta content="text/html; charset=utf-8" http-equiv="Content-Type" /> | ||
| - | + | <script src="http://code.jquery.com/jquery-1.9.1.min.js"></script> | |
| + | <script src="https://2014hs.igem.org/Team:CSWProteens/jquery.slides.min?action=raw&ctype=text/javascript"></script> | ||
<style type="text/css"> | <style type="text/css"> | ||
| + | .auto-style1 { | ||
| + | font-family: Arial; | ||
| + | font-size: 12pt; | ||
| + | margin-top: 0px; | ||
| + | margin-bottom: 0px; | ||
| + | margin-right: 0px; | ||
| + | margin-left: 0px; | ||
| + | color: #000000 | ||
| + | } | ||
| + | |||
.auto-style2 { | .auto-style2 { | ||
| - | + | text-align: center; | |
} | } | ||
| + | |||
.auto-style3 { | .auto-style3 { | ||
| - | font-size: | + | text-align: center; |
| - | + | font-family: Arial; | |
| + | font-size: 50pt; | ||
| + | margin-top: 26px; | ||
| + | margin-bottom: 0px; | ||
| + | color: #058C1D; | ||
} | } | ||
| + | |||
.auto-style4 { | .auto-style4 { | ||
| - | color: #058C1D; | + | text-align: center; |
| + | font-family: Arial; | ||
| + | font-size: 19pt; | ||
| + | margin-top: 0px; | ||
| + | margin-bottom: 0px; | ||
| + | color: #058C1D; | ||
} | } | ||
| - | + | ||
| - | + | .auto-style5 { | |
| + | margin-top: 0px | ||
} | } | ||
| - | + | ||
| - | color: # | + | .auto-style6 { |
| + | font-family: Arial; | ||
| + | font-size: 20pt; | ||
| + | margin-top: 10px; | ||
| + | margin-bottom: 10px; | ||
| + | color: #000000; | ||
| + | |||
| + | .auto-style7 { | ||
| + | font-family: Arial; | ||
| + | font-size: 20pt; | ||
| + | margin-top: 5px; | ||
| + | margin-bottom: 5px; | ||
| + | margin-right: 0px; | ||
| + | margin-left: 0px; | ||
} | } | ||
| - | + | ||
| - | + | #content{ | |
| + | background-image:url(http://upload.wikimedia.org/wikipedia/commons/a/ac/White_flag_of_surrender.svg); | ||
} | } | ||
| - | . | + | |
| - | + | ||
| + | </style> | ||
| + | |||
| + | <style type="text/css"> | ||
| + | @import url(http://fonts.googleapis.com/css?family=Arial); | ||
| + | ul, li { | ||
| + | z-index:999; | ||
| + | margin: 0; | ||
| + | padding: 0; | ||
} | } | ||
| - | + | ||
| - | + | #nav li > a { | |
| - | + | transition: all 0.2s ease-in-out; | |
| - | + | ||
| - | + | ||
| - | + | ||
} | } | ||
| - | + | ||
| - | + | #nav { | |
| - | + | -webkit-box-shadow: 0px 0px 5px 3px rgba(0, 0, 0, 0.75); | |
| - | + | -moz-box-shadow: 0px 0px 5px 3px rgba(0, 0, 0, 0.75); | |
| - | + | box-shadow: 0px 0px 5px 3px rgba(0, 0, 75, 0.75); | |
| - | + | font-family: 'Dosis', sans-serif; | |
| - | + | font-size: 24px; | |
| + | text-align:center; | ||
| + | background-color: #ffffff; | ||
} | } | ||
| - | + | #nav a { | |
| - | + | color: #000000; | |
| - | + | text-decoration: none; | |
} | } | ||
| - | + | #nav li { | |
| - | + | display: inline; | |
| - | + | position: relative; | |
| - | + | ||
| - | + | ||
} | } | ||
| - | + | #nav li ul { | |
| - | + | position: absolute; | |
| - | + | left: -9999px; | |
} | } | ||
| - | + | #nav li a { | |
| - | + | display: inline-block; | |
| - | + | padding: 20px; | |
} | } | ||
| - | + | #nav li:hover a { | |
| - | + | background-color: #000000; | |
| - | + | color: #ffffff; | |
| - | + | ||
| - | + | ||
| - | + | ||
} | } | ||
| - | + | #nav li:hover ul { | |
| - | + | left: 0; | |
| - | + | text-align: left; | |
| - | + | width: 200px; | |
| - | + | box-shadow: 0px 1px 1px #20d681; | |
| - | + | ||
} | } | ||
| - | + | #nav li:hover ul a { | |
| - | + | display: block; | |
| + | padding: 15px 30px 15px 30px; | ||
| + | border-bottom: 1px solid #70a667; | ||
} | } | ||
| - | # | + | #nav li:hover ul a:hover { |
| - | background- | + | background-color: #70a667; |
| + | color: #40004b; | ||
} | } | ||
</style> | </style> | ||
| Line 86: | Line 130: | ||
<meta content="revealTrans(Duration=3.0,Transition=2)" http-equiv="Site-Enter" /> | <meta content="revealTrans(Duration=3.0,Transition=2)" http-equiv="Site-Enter" /> | ||
</head> | </head> | ||
| + | <center><a href="https://2014hs.igem.org/Team:CSWProteens/project"><img alt="" class="auto-style11" height="200" src="https://static.igem.org/mediawiki/2014hs/c/cd/Plantifreeze.gif" width="200" /><img src="https://static.igem.org/mediawiki/2014hs/8/87/Banner.png" width="500"> | ||
| + | </a> | ||
| - | < | + | <a href="https://2014hs.igem.org/Main_Page"><img alt="" class="auto-style5" height="200" src="https://static.igem.org/mediawiki/2014hs/2/26/Igem_grpyon.png" width="200" /></a> </p></center> |
| - | < | + | <body style=" color: #FFFFFF;background-color: #000000; |
| - | + | background-image: url('http://upload.wikimedia.org/wikipedia/commons/a/ac/White_flag_of_surrender.svg');"> | |
| - | </ | + | <ul id="nav"> |
| - | </ | + | <li><a href="https://2014hs.igem.org/Team:CSWProteens"><b>Home</b></a></li> |
| - | + | <li> | |
| - | </ | + | <a href="https://2014hs.igem.org/Team:CSWProteens/team"><b>Team</b></a> |
| - | < | + | <ul> |
| - | </ | + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/team">Members</a></li> |
| + | <li><a href="http://www.csw.org/">The Cambridge School of Weston</a></li> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/advisors">Advisors</a></li> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/sponsors">Sponsors</a></li> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/Attributions">Attributions</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="https://2014hs.igem.org/Team:CSWProteens/project"><b>Project</b></a> | ||
| + | <ul> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/project/frostprotection">Frost Protection</a></li> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/project">Project Description</a></li> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/project/Methods">Methods</a></li> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/project/results">Results</a></li> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/project/discussion">Discussion</a></li> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/project/safety">Safety</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="https://2014hs.igem.org/Team:CSWProteens/notebook"><b>Notebook</b></a> | ||
| + | <ul> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/notebook/">Lab Journal</a></li> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/notebook/protocols">Protocols</a></li> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/notebook/parts">Parts</a></li> | ||
| + | <li><a href="https://2014hs.igem.org/Team:CSWProteens/notebook/references">References</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="https://2014hs.igem.org/Team:CSWProteens/humanpractices"><b>Outreach</b></a> | ||
| + | <ul> | ||
| + | |||
| + | </ul> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="https://2014hs.igem.org/Team:CSWProteens/contact"><b>Contact Us</b></a> | ||
| + | <ul> | ||
| + | <li><a href="http://csw-igem.tumblr.com">Tumblr</a></li> | ||
| + | <li><a href="http://www.facebook.com/cswigem">Facebook</a></li> | ||
| + | <li><a href="mailto:hgoldsweig@csw.org?subject=iGEM">Email</a></li> | ||
| + | </ul> | ||
| + | </ul> | ||
| + | <p> | ||
| + | <center><p class="auto-style6"><b>L A B N O T E B O O K:</b><p></p><p class="auto-style4"> | ||
| + | 2013: <a href="#nov"> November </a> | <a href="#dec"> December </a> | 2014: <a href="#jan"> January </a> | <a href="#feb"> February </a> | <a href="#apr"> April </a> | <a href="#may"> May </a> | <a href="#jun"> June </a></p> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2014hs/7/7a/Tumblr_myr5o6C8Fj1t12eweo2_1280.jpg" width="600"> | ||
| - | < | + | <p><p> |
| + | <center><p class="auto-style4"><b> | ||
| + | <a id="nov">November 13</a></b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | Our first full-length meeting of the year. We spent much of the time discussing the logistical (financial, organizational, etc.) details of the iGEM Competition, before splitting off to confer with our small, self-assigned subcommittees: Design, Communications, Fundraising, and Project Management. | ||
| + | <p><img src="https://static.igem.org/mediawiki/2014hs/0/0b/Tumblr_mwfxda1gL81t12eweo1_1280.jpg" width="400"><br><i>Pictured: <b>Howard</b>, a co-teacher and Faculty Advisor, presenting on the necessary functions of a cohesive iGEM team.</i> | ||
| - | <p class="auto- | + | <p> |
| - | November</ | + | <center><p class="auto-style4"><b> |
| - | <p class="auto- | + | November 18</b></center><p></p> |
| - | < | + | <p class="auto-style1"> |
| - | + | Our lecture half of the class consisted mostly of a detailed overview of synthetic biology, its engineering aspects, and our place in the process. During the lab half, we learned how to make gels for electrophoresis.<p><img src="https://static.igem.org/mediawiki/2014hs/f/fd/Screen_Shot_2014-06-18_at_15.26.28.png" width="400"><br><i>Pictured: Top: <b>Howard</b> draws a diagram of the parts of a biological device.<br> | |
| - | < | + | Bottom: a buffer is added to the gel after it sets.</i> |
| - | < | + | |
| - | + | ||
| - | </ | + | |
<p> | <p> | ||
| + | <center><p class="auto-style4"><b> | ||
| + | November 20</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | We spent the first part of class tossing around ideas for a team name; when it came down to either sticking with the school’s mascot (the gryphon) or choosing something more original, a narrow majority vote decided on the latter. Thus, we are now officially the CSW ProTeens. We then learned more about genetic devices, and specifically their promoters.<p><img src="https://static.igem.org/mediawiki/2014hs/e/ef/Tumblr_mwsnhvAOmN1t12eweo1_1280.jpg" width="400"><br><i>Pictured: Top: <b>Howard</b> describes the inner workings of the lac operon.</i> | ||
| + | |||
| + | |||
<p> | <p> | ||
| - | <p | + | <center><p class="auto-style4"><b> |
| - | <center>< | + | November 25</b></center><p></p> |
| - | + | <p class="auto-style1"> | |
| + | Today, Joey was in sole charge of the class. His lecture covered plasmid backbones in detail. We also watched this CrashCourse video on transcription and translation. For the lab, we learned (or reviewed) how to make agarose gels, use parafilm, and operate a centrifuge. | ||
| + | <p><img src="https://static.igem.org/mediawiki/2014hs/2/23/Screen_Shot_2014-06-18_at_15.29.38.png" width="400"><br><i>Pictured: Left: Joey illustrates how the backbone of a plasmid can be cut to accept new parts.<br> | ||
| + | Right: Aiden pours a gel as Liam and Mason look on.</i> | ||
| + | <p> | ||
| + | <center><b><p class="auto-style4"><a id="dec"> | ||
| + | December 4</b></a></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | Today’s lecture was on translational control, and the workings of DNA polymerase. For the lab, we began to work on making our own ampicillin plates from scratch. | ||
| + | <p><img src="https://static.igem.org/mediawiki/2014hs/3/3d/Screen_Shot_2014-06-18_at_15.31.24.png" width="400"><br><i>Pictured: Top: Howard draws (and vigorously circles) a ribosome binding site.<br> | ||
| + | Bottom: Howard demonstrates the process of making amp plates.</i> | ||
| + | <p> | ||
| + | <center><p class="auto-style4"><b> | ||
| + | Decmber 9</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | This class was dedicated to finishing our ampicillin plates. We also broke in the new autoclave. | ||
| + | <p><img src="https://static.igem.org/mediawiki/2014hs/9/96/Tumblr_myr38eSKII1t12eweo1_1280.jpg" width="400"><br><i>Pictured: The team intently examines the working autoclave.</i> | ||
| + | <p> | ||
| + | <center><p class="auto-style4"><b> | ||
| + | December 11</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | The team split into smaller groups, and each performed a transformation of pPRL, a purple pigment producing plasmid, into E. coli. | ||
| + | <p><img src="https://static.igem.org/mediawiki/2014hs/e/ec/Screen_Shot_2014-06-18_at_15.33.59.png" width="400"><br><i>Pictured: Top: Nate, Aiden and Liam put their plasmids on ice.<br> | ||
| + | Bottom: The plasmids are put on our ampicillin plates.</i> | ||
| + | <p> | ||
| + | <center><p class="auto-style4"><b> | ||
| + | December 16</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | For the lecture portion of class, we learned about “RNA thermometers,” and then discussed logic gates and used them to analyze some devices. We then viewed the (successful!) results of our pPRL transformation.<p><img src="https://static.igem.org/mediawiki/2014hs/5/58/Screen_Shot_2014-06-18_at_15.35.42.png" width="400"><br><i>Top: Howard explains truth tables.<br> | ||
| + | Bottom: Our E. coli is purple!</i> | ||
| - | |||
| - | |||
| - | |||
<p> | <p> | ||
| + | <center><p class="auto-style4"><b> | ||
| + | December 18</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | Joey gave a lecture on 3A assembly. Our lab was to transform pGRN, a green pigment producing plasmid, into E. coli. | ||
| + | <p><img src="https://static.igem.org/mediawiki/2014hs/7/7c/Screen_Shot_2014-06-18_at_16.05.39.png" width="400"><br><i>Top: Joey details the standardized DNA sequences that allow plasmids to be cut.<br> | ||
| + | Bottom-left: Noa and Jenny work on the transformation lab.<br | ||
| + | Bottom-right: Matt, Micah and Aiden need all the hands they can get to stir the bacteria into the Eppendorf tube of plasmids.</i> | ||
| + | </i> | ||
| + | <p> | ||
| + | <center><p class="auto-style4"><b><a id="jan"> | ||
| + | January 6</b></center></a><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | Happy New Year! We dedicated today to preparing gels for our next lab. | ||
| + | <p><img src="https://static.igem.org/mediawiki/2014hs/c/ce/Tumblr_mzcw1a9MBF1t12eweo1_1280.jpg" width="400"><br><i>Pictured: a comb is inserted into the not-yet-solidified gel to create wells.</i> | ||
| + | <p> | ||
| + | <center><p class="auto-style4"><b> | ||
| + | January 8</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | We began a restriction digest lab, but due to technical difficulties had to postpone until the next class. We then split into committees for a regular update. | ||
| + | <p><img src="https://static.igem.org/mediawiki/2014hs/7/73/Tumblr_mzcxqsbk5y1t12eweo1_1280.jpg" width="400"><br><i>Pictured: Joey and Howard divide reagents.</i> | ||
| + | <p> | ||
| + | <center><p class="auto-style4"><b> | ||
| + | January 13</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | We did some more practice with electrophoresis using lambda DNA. | ||
| + | <p><img src="https://static.igem.org/mediawiki/2014hs/7/72/Screen_Shot_2014-06-18_at_16.12.19.png" width="400"><br><i>Pictured:<br> | ||
| + | Top: Noa removes a gel from the electrophoresis chamber.<br> | ||
| + | Bottom: Our results were highly visible when the gels were placed on a light box.</i> | ||
| + | <p> | ||
| + | <center><p class="auto-style4"><b> | ||
| + | January 15</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | We dedicated the whole meeting to practicing with a restriction digest. | ||
| + | <p><img src="https://static.igem.org/mediawiki/2014hs/5/51/Tumblr_n0nxsme1JJ1t12eweo1_1280.jpg" width="400"><br><i>Pictured: <br>Nate, Aiden, and a blurry Liam fill a gel’s wells.</i> | ||
| + | <p> | ||
| + | <center><p class="auto-style4"><b> | ||
| + | January 22</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | In pairs and singles, we researched and presented the team’s top ten project ideas and began to narrow down that list. | ||
| + | <p> | ||
| + | We dedicated the whole meeting to practicing with a restriction digest. | ||
| + | <p><img src="https://static.igem.org/mediawiki/2014hs/8/8d/Screen_Shot_2014-06-18_at_16.14.14.png" width="400"><br><i>Pictured:<br> | ||
| + | Top: Liam and Aiden present their findings.<br> | ||
| + | Bottom: Noa and Jenny set up before sharing theirs.</i> | ||
| + | <p> | ||
| + | <center><p class="auto-style4"><b> | ||
| + | January 27</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | After the previous week’s presentations, we worked to more fully develop the most feasible ideas and draft circuits for them. | ||
| + | <p><img src="https://static.igem.org/mediawiki/2014hs/5/54/Screen_Shot_2014-06-18_at_16.15.48.png" width="400"><br><i>Pictured:<br> | ||
| + | Top: Joey and Howard review the process of writing a circuit.<br> | ||
| + | Bottom: Lilly begins to draft a circuit on the whiteboard.</i> | ||
| - | <center><b> | + | <p> |
| - | We | + | <center><p class="auto-style4"><b> |
| - | We | + | January 29</b></center><p></p> |
| + | <p class="auto-style1"> | ||
| + | We gave our final presentations on the most feasible project ideas, which we had been working on for weeks. | ||
| + | <p><img src="https://static.igem.org/mediawiki/2014hs/0/01/Screen_Shot_2014-06-18_at_16.17.32.png" width="400"><br><i>Pictured:<br> | ||
| + | Top: Summer discusses her proposed circuit.<br> | ||
| + | Bottom: The final list of ideas to be discussed and voted on. (Voting results coming soon…)</i> | ||
| + | <p> | ||
| + | <center><p class="auto-style4"><b><a id="feb"> | ||
| + | February 3</b></center></a><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | The results are in! We, the CSW ProTeens, will pursue plantifreeze for our final project. "Plantifreeze" is our shorthand name for a bacteria that prevents frost from forming on fruits near the end of the fall growing season—in other words, a plant antifreeze | ||
| + | <p><img src="https://static.igem.org/mediawiki/2014hs/d/d4/Tumblr_n0o1ibxaQD1t12eweo1_1280.jpg" width="400"><br><i>Pictured: Liam reviewing his proposed circuit for the project, just before the team splits into smaller groups to research various parts.</i> | ||
| - | <center><b> | + | <center><p class="auto-style4"><b>March Planning</b></center> |
| - | + | <p></p> | |
| - | + | <p class="auto-style1"> | |
| + | Our school has a schedule where courses run for sections of 5-weeks. After February, our class section finished and we did not have a structured meeting time. In this interim, we worked on planning out our project, contacting advisors, and ordering parts for our project. | ||
| + | <p> | ||
| + | <center><p class="auto-style4"><b><a id="apr"> | ||
| + | April 27</b></center></a><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | This weekend we made stock solutions of antibiotic solutions, prepared sterile LB agar plates supplemented with ampicillin, chloramphenicol, and the combination. We also prepared SOB broth for growing up transformed bacteria. Consequently, we are ready now to begin assembly of our device. | ||
| - | |||
| - | |||
| - | |||
<p> | <p> | ||
| + | <center><p class="auto-style4"><b> | ||
| + | April 28</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | We received the pGL575 plasmid from Utah State today by FedEx. Asif suggested that we transform our competent cells with the plasmid ASAP. So we did just that. The plasmid carries the gene for chloramphenicol resistance so we used LB agar with chloramphenicol to select for the transformed cell. We made 5 agar plates so we should have plenty of the plasmid. This plasmid contains the genes for allowing the secretion of proteins made by the bacteria via Type 2 secretion system. Our other plasmid that we are assembling in the lab, regulates secretion of the RiAFP protein. That's why our device is a dual plasmid system: one plasmid controls expression of the protein and the other plasmid regulates secretion | ||
| - | <center><b> | + | <p> |
| - | + | <center><p class="auto-style4"><b> | |
| + | April 29</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | We just checked the selection plates for transformed NEB 10 beta cells. Four of the five plates showed colonies so we were successful with the transformation. The plates are in the frig ready to be grown up to supply the pGL575 plasmid for the device. Next step is to assemble the gene for expression of RiAFP with tag for 6xHis and peptide signal for secretion. We need to put that fusion gene into a plasmid | ||
| - | |||
| - | |||
<p> | <p> | ||
| + | <center><p class="auto-style4"><b><a id="may"> | ||
| + | May 1</b></center></a><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | Kudos to Aiden who did the majority of the lab work. We took the gBlocks of parts A and B of the expression plasmid and the linearized plasmid pSB3A1 and digested the prefix and suffix of each with restriction enzymes: EchoRI, SpeI, Xbal, PstI, PslI. These enzymes recognize restriction sites and cut them in ways that allow part A to fuse with part B and for the fusion of A-B to fit into pSB3A1. The next step is to permanently ligate the parts so that we end up with a circular plasmid carrying the fusion part. The ligation will be performed on Thursday morning between 10 am and 1 pm. Drop by. | ||
| - | |||
| - | |||
| - | |||
<p> | <p> | ||
| + | <center><p class="auto-style4"><b> | ||
| + | May 6</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | The transformation failed. We later discovered that we used 0.2 micrograms of plasmid instead of the suggested 1 microgram while constructing the expression plasmid from parts A and B. This altered the volume of the reaction, and was likely the cause of failure. | ||
| - | |||
| - | |||
| - | |||
<p> | <p> | ||
| + | <center><p class="auto-style4"><b> | ||
| + | May 8</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | In absence of gBlocks, we can not do anything with the expression plasmid. We took individual colonies of NEB 10 cells transformed with pLG575 plasmids and put them in LB broth cultures to incubate at room temp for 24 hours. Growing up these cells will afford us a large supply of the pLG575 plasmids. | ||
| + | <p> | ||
| + | We ordered more gBlocks from IDT. We will carefully rewrite the protocol for digestion and ligation of the parts that go into the linearized pSB3A1 so transformation can be successful with a future attempt. Also will run concurrent positive control in the future to avoid our inability to analyze any future failures. | ||
| - | + | <p> | |
| - | + | <center><p class="auto-style4"><b> | |
| + | May 14</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | After very careful preparation, the parts for the expression plasmid were digested, ligated and the resulting ligation product was used to transform NEB 10 cells today with the capable assistance of Aiden, Nate, and Noa. Many things were changed from the unsuccessful transformation from last week. The transformed cells are in the 37 degree C incubator so please everyone hold your breath | ||
| - | <center><b> | + | <p> |
| - | + | <center><p class="auto-style4"><b> | |
| + | May 15</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | Transformation is successful. Next step is to grow out colonies of transformed cell in liquid culture so we have good supply. Then we mini prep both plasmids and cotransform DB21 DE3 cells and look for recombinant protein in supernatant and lysate. | ||
| + | <p> | ||
| + | So we have the expression plasmid in NEB 10 beta cells on agar supplemented with amp. The next step is to grow up colonies form the agar plate in LB broth. This will give us a large supply of plasmids. We will miniprep the liquid culture getting bare plasmids for contranformation of DB21 DE3 competent cells with both plasmids. By using Western blot we confirm presence of RiAFP in supernatant and lysate. What did we do right this time. In the ligation step, we were very careful to use the correct proportion of DNA from the inserts (Parts A and B) and the vector (linearized pSB1A3). We used 50 ng of vector DNA and made use of a calculator on the NEB web site to calculate the amt of insert DNA to use. There should be a 3 to 1 molar excess of insert DNA to vector DNA to ensure that all the linearized vector is fully occupied by insert DNA. The calculation used the nucleotide lengths of the inserts (about 450 bp for each part) and the vector (2155 bp) to make the calculation. Also in an incubation in ligation we incorrectly used 22 degrees C instead of room temperature originally. This was corrected in the lates transformation. | ||
| - | <center><b> | + | <p> |
| - | + | <center><p class="auto-style4"><b> | |
| + | May 18</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | After we transformed the NEB 10 beta cells with the newly assembled plasmid (T7 RBS RiAFP 6xHis HlyA term term), Ellie selected a few individual colonies from the agar plate supplemented with amp and put them in 50 mL of LB broth supplemented with amp in sterile 250 ml flasks with a sterile magnetic stirrer and grew up the cells over 16 hours at room temperature with gentle stirring. Our newly acquired rotating shaker has a broken belt so it is on the bench. After the 16 hour incubation, the broth had turned from clear to opaque. We transferred 10 ml of the broth to sterile falcon tubes and stored them at 4 degrees C. We do minipreps on the cells to isloate and pruify the plasmids. We also transformed NEB 10 cells wit the pLG575 plasmids and stored them in10 ml broth at 4 degrees C. So now we have ample supplies of both plasmids. | ||
| + | <p> | ||
| + | <p class="auto-style1"> | ||
| + | The next step is to miniprep the transformed cells. Then we cotransform the two plasmids into DB21 DE3 competent cells. Our device includes an expression vector containing RiAFP cloned downstream of the T7 promoter. DB21 DE3 competent cells carry a chromosomal copy of the phage T7 RNA Polymerase gene, which is controlled by a lac promoter. When inducer (IPTG) is added, T7 RNA Polymerase is expressed and becomes dedicated to transcription of RiAFP. | ||
| + | <p> | ||
| + | <p class="auto-style1"> | ||
| + | Plasmid pLG575 includes the coding regions for proteins HlyB and HlyD. | ||
| - | |||
| - | |||
<p> | <p> | ||
| + | <center><p class="auto-style4"><b> | ||
| + | May 20</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | We are ordering the DB21 DE3 competent cells from NEB very shortly in preparation of cotransformation of these cells with the expression and secretion plasmids. | ||
| - | |||
| - | |||
<p> | <p> | ||
| + | <center><p class="auto-style4"><b> | ||
| + | May 22</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | We took individual colonies growing on agar supplemented with chloramphenicol and put them in SOB culture broth. SOB is a slightly modified LB broth. The cells, NEB 10 beta cells, transformed with pLG575 plasmid will grow in the broth and provide a source of the plasmid. We also transformed NEB 10 cells with the expression plasmid. | ||
| + | <p> | ||
| + | <p class="auto-style1"> | ||
| + | Currently we have liquid cultures of NEB cells with the expression plasmid and liquid cultures of NEB cells with the secretion plasmid. At this point we are poised to do minipreps on both types of cultures. | ||
| + | <p> | ||
| + | <p class="auto-style1"> | ||
| + | The minipreps will give us bare plasmids that in turn will be used to transform BL21 DE3 E coli cells. These are chemically competent E. coli cells suitable for transformation and T7 promoter driven expression of RiAFP expression. The two plasmids will be used to cotransform BL21 DE3 cells. Following cotransformation, expression of RiAFP will be induced by IPTG. | ||
| - | <center><b> | + | <p> |
| - | + | <center><p class="auto-style4"><b> | |
| + | May 24</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | On Tuesday we will do miniprep on the cells containing the expression plasmid. At that point we will be in a position to cotransform the BL21 DE3 cells that constitute the PlantiFreeze device. Competent BL21 DE3 cells will be shipped to the lab on Wednesday on dry ice. We will transform the cells on Wednesday. On Thursday the 16 hour incubation of the transformed cells will be ready. On Thursday, we will take individual colonies from the agar plate and put them into SOC media. On Friday of next week if all goes well we will have an ample supply of the BL21 DE3 cells transformed with both the expression and secretion plasmids. At this point the moment of true has arrived! By introducing IPTG into the culture we should be able to induce the cells to express RiAFP and secrete the recombinant protein into the medium in which the cells are growing. Here is where Steph and the Silver lab come into action. We need to assay the medium and the lyzed cells to confirm the RiAFP was expressed by the E coli and secreted. We will do this using Western blot. | ||
| + | <p> | ||
| + | <p class="auto-style1"> | ||
| + | There will be other analyzes that we'll also do:<br> | ||
| + | • Effect of each plasmid and both plasmids on the growth rate of BL21 DE3 cells (by turbidity analysis using spectrophotmometer<br> | ||
| + | • Gel electrophoresis of some of the intermediate constructs | ||
| - | <center><b> | + | <p> |
| - | + | <center><p class="auto-style4"><b> | |
| + | May 24</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | Rather than using Western blot we are thinking about the use of Invision In-gel stain from Life Technologies. Visualization of the fluorescent bands uses: | ||
| + | <p> | ||
| + | <p class="auto-style1"> | ||
| + | UV transilluminator (302 nm) equipped with a camera capable of integration<br> | ||
| + | To view and photograph a gel on the UV transilluminator, use a video camera, CCD (Charged Couple Device) camera, or a cooled CCD camera with ethidium bromide filter or band pass filter encompassing the emission maxima | ||
| + | (590 nm) of the stain. | ||
| - | <center><b> | + | <p> |
| - | + | <center><p class="auto-style4"><b> | |
| - | " | + | May 27</b></center><p></p> |
| + | <p class="auto-style1"> | ||
| + | DL21 BE3 competent cells from NEB arrive tomorrow. Today we will do miniprep on the cells containing the expression plasmid. <br> | ||
| + | So how 'bout we order:<br> | ||
| + | InVision™ His-Tag In-Gel Staining Kit<br> | ||
| + | InVision™ His-tag In-gel Stain is supplied as a 1X ready-to-use staining reagent (500 mL) [enough to run 20 mini gels]<br> | ||
| + | BenchMark™ His-tagged Protein Standard (125 µL) is also included in the kit<br> | ||
| + | NuPAGE® Novex® 10% Bis-Tris Gels (pre-cast polyacrylamide gels)<br> | ||
| - | <center><b> | + | <p> |
| - | This | + | <center><p class="auto-style4"><b> |
| - | + | May 28</b></center><p></p> | |
| + | <p class="auto-style1"> | ||
| + | Thanks to Joey and Thomas for doing the miniprep on the expression plasmid. We now have pure DNA from both plasmids and are poised to cotransform BL21 DE3 competent cells. This is the last step in the assembly of the PlantiFreeze device. After that step we do functional assays to confirm that the device works to express and secret RiAFP. As a side venture we'll be exploring the effect of transformation of the BL23 DE 3 cells with each plasmid alone and both together. We'll be constructing growth curves based on optical density of the culture media. | ||
| + | <p> | ||
| + | <p class="auto-style1"> | ||
| + | We have ordered from LifeTechnologies Invision in-gel stain, a latter, and precast gels for confirming the expression and secretion of the protein. These experiments will be done with Steph Hayes at Pam Silver's lab at Harvard Medical School. | ||
| + | <p> | ||
| + | <p class="auto-style1"> | ||
| + | Keep your fingers crossed that everything goes right and we'll be in the home stretch. Still need to do data analysis, the Wiki, our poster and the presentation so we need everybody at battle stations a while longer. | ||
| + | <p> | ||
| + | <p class="auto-style1"> | ||
| + | Finally, we need to do more minipreps more growing up liquid cultures, more transformations so please avail yourself of the opportunity to get your hands wet in the lab. Its fun, its instructional, and you help is really neede | ||
| - | <center><b> | + | <p> |
| - | + | <center><p class="auto-style4"><b> | |
| + | May 29</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | Lab today 1 to 5:<p> | ||
| + | <p class="auto-style1"> | ||
| - | < | + | • cotransformation of Bl21 De 3 cells<br> |
| - | + | • miniprep of plasmids<br> | |
| + | • making agar plates<br> | ||
| + | • making liquid cultures<br> | ||
| + | • planning measurement of oD of culture for growth curves | ||
| - | <center><b> | + | <p> |
| - | Yesterday | + | <center><p class="auto-style4"><b> |
| + | May 31</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | Yesterday was a banner day for lab work. Thanks to Joey, Micah, Aiden, and Thomas. We did a miniprep of the NEB 10 cells transformed with the secretion plasmid pLG575. Did the milestone cotransformation of BL21 DE3 cells with the expression and secretion plasmids. On Saturday after 16 hours incubation at 37 degrees C we should know if it worked. This time we ran a positive control of pUC16 DNA so if something goes wrong will have the control to help us unravel the problem. | ||
| + | <p> | ||
| + | <p class="auto-style1"> | ||
| + | If the cotransformation works our next step is to grow up the cotransformed cells in LB broth and then induce them with IPTG to express and secrete RiAFP. | ||
| + | <p> | ||
| + | <p class="auto-style1"> | ||
| + | Remember we plan on determining the influence on, the other and both plasmids on the growth of BL21 DE3 cells using the optical density of the liquid culture on the spectrophotometer. If time before the close of the Wiki permits we might also do digestions of the plasmids and look at their electrophoresis patterns. | ||
| - | <center><b> | + | <p> |
| - | + | <center><p class="auto-style4"><b><a id="jun"> | |
| + | June 1</b></center><p></a></p> | ||
| + | <p class="auto-style1"> | ||
| + | No growth on either the plates with contransformation or the positive control. It is obviously useful to have a positive control because now we can conclude that we are doing something wrong with the transformation procedure or the competent cells are no longer competent. In fact, in our hands, tansformation has been very unreliable. But this is the first time we ran a positive control. | ||
| - | <center><b> | + | <p> |
| - | The | + | <center><p class="auto-style4"><b> |
| + | June 2</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | The NanoDrop is a specialized spectrophotometer that measures the amount of DNA in a sample as small as 1 microliter. | ||
| + | <p> | ||
| + | <p class="auto-style1"> | ||
| + | We are having trouble with the cotransformation and we are worried that the plasmid DNA we are getting from the minipreps was too little. As a benchmark, it is good to have more than 50 nanograms/microliter of plasmid DNA. | ||
| + | <p> | ||
| + | <p class="auto-style1"> | ||
| + | We have two plasmids: pLG575 and the expression plasmid. We have 5 nanograms/microliter of this plasmid. Obviously very low. The other plasmid is the expression plasmid. We have 47 nanograms/microliter of this plasmid. Good. | ||
| + | <p> | ||
| + | <p class="auto-style1"> | ||
| + | We can adjust the miniprep to increase the concentration of pLG575 DNA, | ||
| + | <p> | ||
| + | <p class="auto-style1"> | ||
| + | The puzzle is that we should have gotten transformation of the expression plasmid. So we still have to look at the transformation procedure. | ||
| - | <center><b> | + | <p> |
| - | + | <center><p class="auto-style4"><b> | |
| + | June 6</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | This has been a frustrating and puzzling week. The cotransformation performed on Sunday failed, leading us to wonder if we got plasmid DNA from the minipreps of the expression and secretion plasmids. Steph generously allowed us to use the Nanodrop analyzer in her lab. We found that the DNA concentration was low but acceptable for pLG575 and in a good range for the expression plasmid. We made some alterations to the miniprep procedure. We repeated the transformations in a wide ranging way and were amazed that the only plate with growth was the one with agar only. This observation directed us to think about a problem with the antibiotics so we made new stock solutions and were especially careful about antibiotic concentrations in the most recent transformations. On Thursday, Joey, Micah, and Thomas were in the lab. We used BL21 DE competent cells and did the following: | ||
| + | <p> | ||
| + | <p class="auto-style1"> | ||
| + | • cotransformation with the original pLG575 plasmid from Utah State and the expression plasmid<br> | ||
| + | • transformation of pUC19 positive control plasmid<br> | ||
| + | • transformation with pLG575 plasmid obtained by miniprep using modifications to increase concentration<br> | ||
| + | • transformation of the pPURP plasmid from the BioBuilder lab exercise<br> | ||
| - | + | All in all, maddeningly frustrating, but we will persevere. | |
| - | + | ||
| - | <center><b> | + | <p> |
| - | + | <center><p class="auto-style4"><b> | |
| + | June 7</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | Puzzling, frustrating, and humiliating! We used commercial BL21 DE3 competent cells in the transformation reactions. pLG575 (orig) refers to the plasmids shipped to use by Utah State; pLG575 (miniprep) refers to the plasmid DNA we got by miniprep of the transformed NEB 10 beta cells transformed with the pLG575 plasmids. | ||
| + | <p><p class="auto-style1"> | ||
| + | pPURP is the plasmid from BioBuilders and pUC19 is the positive control plasmid sent with the competent cells by NEB. | ||
| + | <p><p class="auto-style1"> | ||
| + | <img src="https://static.igem.org/mediawiki/2014hs/0/03/Screen_Shot_2014-06-18_at_15.14.16.png"> | ||
| + | <p> <img src="https://static.igem.org/mediawiki/2014hs/2/24/Screen_Shot_2014-06-18_at_15.14.26.png"> | ||
| + | |||
| + | <p><p class="auto-style1"> | ||
| + | We mixed new stock solution of ampicillin and chloramphenicol and carefully checked the concentrations in the LB agar, so I am really bewildered by the discrepancies between the expected and observed results. | ||
| - | |||
| - | |||
| - | <center><b> | + | <p> |
| - | + | <center><p class="auto-style4"><b> | |
| + | June 10</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | Thank you Aiden, Noa, Ben, and Joey for working in Steph's lab at Harvard Medical School. In an attempt at troubleshooting our problem with transformation we did transformations with several different strains of competent cells. The plasmids we used included: pLG575, the expression plasmid, pUC19. | ||
| - | |||
| - | |||
| - | <center><b> | + | <p> |
| - | + | <center><p class="auto-style4"><b> | |
| + | June 11</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | Yesterday, we used five different strains of competent E coli and five separate plasmids for transformation of the strains of competent cells. Here are the results of the growth of the plates with the appropriate antibiotics: | ||
| - | < | + | <p><p class="auto-style1"><b>Results from Steph’s Lab</b><p> |
| - | + | <img src="https://static.igem.org/mediawiki/2014hs/1/1a/Screen_Shot_2014-06-18_at_15.04.37.png"><p> | |
| + | Disregarding one or two anomalies, the analysis indicates two things: | ||
| + | <p> | ||
| + | <p class="auto-style1"> | ||
| + | The BL21 DE3 competent cells don't appear to be competent. Keeping them on dry ice may not have been able to maintain a stable -80 degree centigrade temp allowing them to become incapable of transformation (see column pUC19). Please note that the prior batch of competent cells from NEB (NEB 10 beta cells) were transformed by the pLG575 plasmids we recieved from Utah State and the plasmid we constructed from the gBlocks form IDT. I did not observe thawing of the BL23 DE3 cells. The plasmid DNA we obtained by miniprep were not able to transform any competent cells (see last three columns). The cause of the failure of the miniprep remains uncertain. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | <center><b> | + | <p> |
| - | + | <center><p class="auto-style4"><b> | |
| + | June 12</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | Hi, all. Let’s summarize where we are at the moment. | ||
| + | <p> | ||
| + | <p class="auto-style1"> | ||
| + | We have plasmids with the expression cassette (amp resistant) and the secretion cassette (chloramphenicol resistant), respectively in transformed NEB 10 beta cells. This is where we reached a stone wall. Using minipreps, we were unable to cotransform BL21 DE3 cells and induce those cells with IPTG to express and secrete RiAFP. We have an in-gel stain that will enable us to detect RiAFP in cytoplasm and culture media standing by and ready to use (takes about 3 hours to do). | ||
| + | <p> | ||
| + | <p class="auto-style1"> | ||
| + | Under the guidance of Steph, team members performed transformations using several E coli strains and different plasmids (the results summarized in an email message sent to all yesterday). The analysis of those results suggest that the commercial competent cells from NEB (BL21 D3) were degrade by unstable maintenance of -80 C temp and that our miniprep procedure was flawed. | ||
| + | <p> | ||
| + | <p class="auto-style1"> | ||
| + | It is easy to overcome the competent cell flaw. Not so easy is pin pointing what's wrong with the miniprep. Since we have amp and chloramphenicol resistant NEB 10 cells in the Bio 3 frig, we can analysis the plasmids in those cells and figure out a strategy to go forward. We have in the frig in Bio 3 agar plates and LB broth cultures of the NEB 10 beta cells which are amp resistant cells andchloramphenicol resistant cells meaning they were transformed by the expression and secretion plasmids, respectively. I will bring them to the AlbertsLab tomorrow for analysis confirming the presence of the appropriate plasmids in the cells. | ||
| + | <p> | ||
| + | <p class="auto-style1"> | ||
| + | Needless to say, the clock is ticking and it will be a challenge to finish all the lab work. The Wiki closes on 20 June, but the lab work can go on beyond that date and any new data can be incorporated in the Poster and the PowerPoint slide deck. Even then time may run out despite the best efforts. | ||
| - | |||
| - | |||
| - | <center><b> | + | <p> |
| - | + | <center><p class="auto-style4"><b> | |
| + | June 13</b></center><p></p> | ||
| + | <p class="auto-style1"> | ||
| + | Noa's review of the Parts synthesized by IDT prompted me to do a deep dive into the parts sequence synthesized by IDT. Oh boy, did we mess up and compounding that problem we didn't check the parts sequence when they came back from IDT. But since they have a 3-5 business day turn around we can recover but we want to order just one DNA fragment which will be inserted into a linearized plasmid with an antibiotic resistance different from the secretion plasmid. That plasmid will then be cotransformed with our secretion plasmid into BL21 DE3 competent cells. Uncovering the incorrect sequence in the expression plasmid solves our current problem of not being able to cotransform in that we'll have a brand new plasmid and we will use new competent cells. | ||
| + | <p><p class="auto-style1"> | ||
| + | Here is new revised approach: | ||
| + | <p><p class="auto-style1"> | ||
| + | we need a plasmid which contains these components and BioBrick standard prefix and suffix (not the RFC 23):<br> | ||
| + | • T7 promoter<br> | ||
| + | • RBS<br> | ||
| + | • RiAFP coding sequence<br> | ||
| + | • HlyA coding sequence<br> | ||
| + | • 6x His marker<br> | ||
| + | • double terminator<p> | ||
| + | <p class="auto-style1"> | ||
| + | We insert (using digestion and ligation) the above fragment into a linearized plasmid with a resistance marker other than chloramphenicol (the secretion plasmid is chloramphenicol resistant). We then use Invision in gel stain to detect presence of RiAFP in cytoplasm and media. | ||
| + | <p> | ||
| + | <p class="auto-style1"> | ||
| + | We'll be working up to the last minute: here's the timeline:<p> | ||
| + | <p class="auto-style1"> | ||
| + | • Receive new part from IDT: 19 June<br> | ||
| + | • Insert new part into linearized plasmid: 19 June<br> | ||
| + | • Transform competent cells to grow up stock of plasmid: 20 June<br> | ||
| + | • Grow up liquid culture of transformant: 21 June<br> | ||
| + | • Miniprep of plasmid DNA: 22 June<br> | ||
| + | • Transform BL21 DE3 cells: 22-23 June<br> | ||
| + | • Grow up transformed BL21 DE3 cells and induce expression and secretion of RiAFP: 23-24 June<br> | ||
| + | • Use Invision system to detect RiAFP in fractions: 25 June<br> | ||
| + | • Analyze results: 25-26 June<p> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | < | + | <p> |
| - | + | <center><p class="auto-style4"><b> | |
| - | + | June 14</b></center><p></p><p class="auto-style1"> | |
| - | + | After the disheartening discovery that Part A had been incorrectly designed, we decided to engineer a single DNA fragment to serve as the expression genetic circuit in thePlantiFreeze Device. The circuit would comprise the following components: | |
| - | + | <p><p class="auto-style1"> | |
| - | + | BioBrick Standard Protein Coding Region Prefix /T7 Promoter /RBS /RiAFP/HlyA /Double terminator /BioBrick Standard Protein Coding Region Suffix | |
| - | + | <p><p class="auto-style1"> | |
| - | + | The DNA sequences for the components could be found in the Registry. So it was not difficult to design the part. It was 864 base pairs in length. It is fast and simple to order a 864 base pair gBlock fragment form IDT (double-stranded, sequence-verified genomic blocks that ship in only a few working days for affordable and easy gene construction). | |
| - | + | <p><p class="auto-style1"> | |
| - | + | The first step in ordering a gBlock is automated review by screening tools and expert review by IDT scientists of entered sequences for characteristics that may interfere with synthesis. This review found that there were sequence repetitions longer than 8 base pairs that comprise a combined 91% of the sequences within a window of 70 base pairs beginning at base pair 235. This structure would interfere with synthesis. So the IDT gene product specialist optimized the sequence for expression in E coli and compatible with the gBlock synthesis process. Optimization theoretically just changes codon usage without changing the proteins coded. Importantly, the EcoRI and PstI sites on the 5’ and 3’ ends of this gene were maintained compatible with BioBrick standard 3A assembly. However, Noa at Steph's lab ran the optimized new sequence thru the seqbuilder program and identified that the optimization process put in another Pst1 restriction site at position 637 in addition to the appropriate one at position 863 (the one compatible with the BioBrick standard). Having the secondPstI restriction site breaks the BioBrick standard and makes correct assembly impossible. Following the finding of this glitch, the fragment was reoptimizedremoving the PstI site at 836. The part was order for synthesis and delivery as soon as possible. | |
| + | <p> | ||
| + | <p><center><a href="https://static.igem.org/mediawiki/2014hs/1/10/IGEM_Seqbuilder_Expression_plasmid_in_backbone.png"><img src="https://static.igem.org/mediawiki/2014hs/1/10/IGEM_Seqbuilder_Expression_plasmid_in_backbone.png" width="450"></a> <a href="https://static.igem.org/mediawiki/2014hs/6/61/Screen_shot_2014-06-13_at_11.04.23_AM.png"> <img src="https://static.igem.org/mediawiki/2014hs/6/61/Screen_shot_2014-06-13_at_11.04.23_AM.png" width="450"></a><br><b>New sequence outlined in SeqBuilder (click to enlarge)</b></center><p class="auto-style1"> | ||
| + | This new part just ordered from IDT would be inserted into a linearized plasmid backbone with antibiotic resistance different from the plasmid controlling secretion (pLG575). We decided to use again pSB1A3. This linearized plasmid is supplied by iGEM. We had exhausted our previous supply so ordered a new one. | ||
| + | <p><p class="auto-style1"> | ||
| + | The plan is to insert the new part into pSB1A3 and again do cotransformation of BL21 DE3 cells with the two plasmids. Then we would induce thecotransformants with IPTG to express and secrete RiAFP. Finally, using theInvision In-gel Staining Kit we would analyze the media and BL21 DE3 cell lyzatefor RiAFP. This would mean we work up to the very last minute before the Jamboree but we are determined to successfully complete the project. | ||
| + | <p><p class="auto-style1"> | ||
| + | One last problem is a reliable source of pLG575. Because our mini-centrifuge has a max speed of 4000 rpm (as compared to the recommended of 12000 rpm) our yield of pLG575 from Qiagen miniprep is abysmal (on Nanodrop: 5 ug/uL) The pLG575 generously sent to us by our advisor at Utah State, Asif Rahman, on Nanodrop showed 260 ug/ul. I'll ask Asif to send us another tube of pLG575 from his lab. Also, we will do the miniprep with Steph using a centrifuge of appropriate speed. | ||
| + | <p><p class="auto-style1"> | ||
| + | So that's our plan! The team is up to the challenge. In the lull while we wait for the new part, we will work on the Wiki due 20 June. and work on the Poster and our 20 minute Presentation. | ||
Latest revision as of 02:19, 21 June 2014



L A B N O T E B O O K:
2013: November | December | 2014: January | February | April | May | June

Our first full-length meeting of the year. We spent much of the time discussing the logistical (financial, organizational, etc.) details of the iGEM Competition, before splitting off to confer with our small, self-assigned subcommittees: Design, Communications, Fundraising, and Project Management.

Pictured: Howard, a co-teacher and Faculty Advisor, presenting on the necessary functions of a cohesive iGEM team.
November 18
Our lecture half of the class consisted mostly of a detailed overview of synthetic biology, its engineering aspects, and our place in the process. During the lab half, we learned how to make gels for electrophoresis.

Pictured: Top: Howard draws a diagram of the parts of a biological device.
Bottom: a buffer is added to the gel after it sets.
November 20
We spent the first part of class tossing around ideas for a team name; when it came down to either sticking with the school’s mascot (the gryphon) or choosing something more original, a narrow majority vote decided on the latter. Thus, we are now officially the CSW ProTeens. We then learned more about genetic devices, and specifically their promoters.

Pictured: Top: Howard describes the inner workings of the lac operon.
November 25
Today, Joey was in sole charge of the class. His lecture covered plasmid backbones in detail. We also watched this CrashCourse video on transcription and translation. For the lab, we learned (or reviewed) how to make agarose gels, use parafilm, and operate a centrifuge.

Pictured: Left: Joey illustrates how the backbone of a plasmid can be cut to accept new parts.
Right: Aiden pours a gel as Liam and Mason look on.
Today’s lecture was on translational control, and the workings of DNA polymerase. For the lab, we began to work on making our own ampicillin plates from scratch.

Pictured: Top: Howard draws (and vigorously circles) a ribosome binding site.
Bottom: Howard demonstrates the process of making amp plates.
Decmber 9
This class was dedicated to finishing our ampicillin plates. We also broke in the new autoclave.

Pictured: The team intently examines the working autoclave.
December 11
The team split into smaller groups, and each performed a transformation of pPRL, a purple pigment producing plasmid, into E. coli.

Pictured: Top: Nate, Aiden and Liam put their plasmids on ice.
Bottom: The plasmids are put on our ampicillin plates.
December 16
For the lecture portion of class, we learned about “RNA thermometers,” and then discussed logic gates and used them to analyze some devices. We then viewed the (successful!) results of our pPRL transformation.

Top: Howard explains truth tables.
Bottom: Our E. coli is purple!
December 18
Joey gave a lecture on 3A assembly. Our lab was to transform pGRN, a green pigment producing plasmid, into E. coli.

Top: Joey details the standardized DNA sequences that allow plasmids to be cut.
Bottom-left: Noa and Jenny work on the transformation lab.
Happy New Year! We dedicated today to preparing gels for our next lab.

Pictured: a comb is inserted into the not-yet-solidified gel to create wells.
January 8
We began a restriction digest lab, but due to technical difficulties had to postpone until the next class. We then split into committees for a regular update.

Pictured: Joey and Howard divide reagents.
January 13
We did some more practice with electrophoresis using lambda DNA.
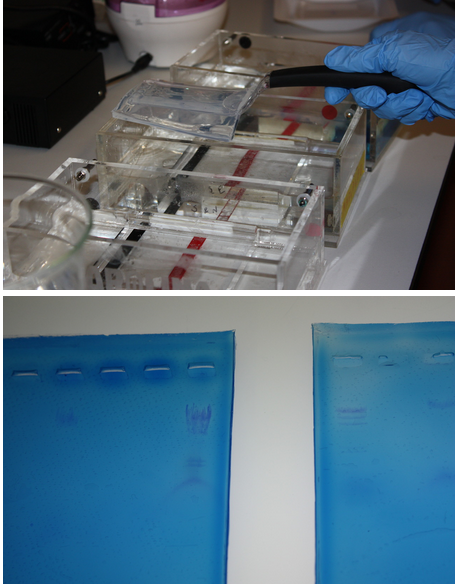
Pictured:
Top: Noa removes a gel from the electrophoresis chamber.
Bottom: Our results were highly visible when the gels were placed on a light box.
January 15
We dedicated the whole meeting to practicing with a restriction digest.

Pictured:
Nate, Aiden, and a blurry Liam fill a gel’s wells.
January 22
In pairs and singles, we researched and presented the team’s top ten project ideas and began to narrow down that list.
We dedicated the whole meeting to practicing with a restriction digest.

Pictured:
Top: Liam and Aiden present their findings.
Bottom: Noa and Jenny set up before sharing theirs.
January 27
After the previous week’s presentations, we worked to more fully develop the most feasible ideas and draft circuits for them.

Pictured:
Top: Joey and Howard review the process of writing a circuit.
Bottom: Lilly begins to draft a circuit on the whiteboard.
January 29
We gave our final presentations on the most feasible project ideas, which we had been working on for weeks.

Pictured:
Top: Summer discusses her proposed circuit.
Bottom: The final list of ideas to be discussed and voted on. (Voting results coming soon…)
The results are in! We, the CSW ProTeens, will pursue plantifreeze for our final project. "Plantifreeze" is our shorthand name for a bacteria that prevents frost from forming on fruits near the end of the fall growing season—in other words, a plant antifreeze

Pictured: Liam reviewing his proposed circuit for the project, just before the team splits into smaller groups to research various parts.
March Planning
Our school has a schedule where courses run for sections of 5-weeks. After February, our class section finished and we did not have a structured meeting time. In this interim, we worked on planning out our project, contacting advisors, and ordering parts for our project.
This weekend we made stock solutions of antibiotic solutions, prepared sterile LB agar plates supplemented with ampicillin, chloramphenicol, and the combination. We also prepared SOB broth for growing up transformed bacteria. Consequently, we are ready now to begin assembly of our device.
April 28
We received the pGL575 plasmid from Utah State today by FedEx. Asif suggested that we transform our competent cells with the plasmid ASAP. So we did just that. The plasmid carries the gene for chloramphenicol resistance so we used LB agar with chloramphenicol to select for the transformed cell. We made 5 agar plates so we should have plenty of the plasmid. This plasmid contains the genes for allowing the secretion of proteins made by the bacteria via Type 2 secretion system. Our other plasmid that we are assembling in the lab, regulates secretion of the RiAFP protein. That's why our device is a dual plasmid system: one plasmid controls expression of the protein and the other plasmid regulates secretion
April 29
We just checked the selection plates for transformed NEB 10 beta cells. Four of the five plates showed colonies so we were successful with the transformation. The plates are in the frig ready to be grown up to supply the pGL575 plasmid for the device. Next step is to assemble the gene for expression of RiAFP with tag for 6xHis and peptide signal for secretion. We need to put that fusion gene into a plasmid
Kudos to Aiden who did the majority of the lab work. We took the gBlocks of parts A and B of the expression plasmid and the linearized plasmid pSB3A1 and digested the prefix and suffix of each with restriction enzymes: EchoRI, SpeI, Xbal, PstI, PslI. These enzymes recognize restriction sites and cut them in ways that allow part A to fuse with part B and for the fusion of A-B to fit into pSB3A1. The next step is to permanently ligate the parts so that we end up with a circular plasmid carrying the fusion part. The ligation will be performed on Thursday morning between 10 am and 1 pm. Drop by.
May 6
The transformation failed. We later discovered that we used 0.2 micrograms of plasmid instead of the suggested 1 microgram while constructing the expression plasmid from parts A and B. This altered the volume of the reaction, and was likely the cause of failure.
May 8
In absence of gBlocks, we can not do anything with the expression plasmid. We took individual colonies of NEB 10 cells transformed with pLG575 plasmids and put them in LB broth cultures to incubate at room temp for 24 hours. Growing up these cells will afford us a large supply of the pLG575 plasmids.
We ordered more gBlocks from IDT. We will carefully rewrite the protocol for digestion and ligation of the parts that go into the linearized pSB3A1 so transformation can be successful with a future attempt. Also will run concurrent positive control in the future to avoid our inability to analyze any future failures.
May 14
After very careful preparation, the parts for the expression plasmid were digested, ligated and the resulting ligation product was used to transform NEB 10 cells today with the capable assistance of Aiden, Nate, and Noa. Many things were changed from the unsuccessful transformation from last week. The transformed cells are in the 37 degree C incubator so please everyone hold your breath
May 15
Transformation is successful. Next step is to grow out colonies of transformed cell in liquid culture so we have good supply. Then we mini prep both plasmids and cotransform DB21 DE3 cells and look for recombinant protein in supernatant and lysate.
So we have the expression plasmid in NEB 10 beta cells on agar supplemented with amp. The next step is to grow up colonies form the agar plate in LB broth. This will give us a large supply of plasmids. We will miniprep the liquid culture getting bare plasmids for contranformation of DB21 DE3 competent cells with both plasmids. By using Western blot we confirm presence of RiAFP in supernatant and lysate. What did we do right this time. In the ligation step, we were very careful to use the correct proportion of DNA from the inserts (Parts A and B) and the vector (linearized pSB1A3). We used 50 ng of vector DNA and made use of a calculator on the NEB web site to calculate the amt of insert DNA to use. There should be a 3 to 1 molar excess of insert DNA to vector DNA to ensure that all the linearized vector is fully occupied by insert DNA. The calculation used the nucleotide lengths of the inserts (about 450 bp for each part) and the vector (2155 bp) to make the calculation. Also in an incubation in ligation we incorrectly used 22 degrees C instead of room temperature originally. This was corrected in the lates transformation.
May 18
After we transformed the NEB 10 beta cells with the newly assembled plasmid (T7 RBS RiAFP 6xHis HlyA term term), Ellie selected a few individual colonies from the agar plate supplemented with amp and put them in 50 mL of LB broth supplemented with amp in sterile 250 ml flasks with a sterile magnetic stirrer and grew up the cells over 16 hours at room temperature with gentle stirring. Our newly acquired rotating shaker has a broken belt so it is on the bench. After the 16 hour incubation, the broth had turned from clear to opaque. We transferred 10 ml of the broth to sterile falcon tubes and stored them at 4 degrees C. We do minipreps on the cells to isloate and pruify the plasmids. We also transformed NEB 10 cells wit the pLG575 plasmids and stored them in10 ml broth at 4 degrees C. So now we have ample supplies of both plasmids.
The next step is to miniprep the transformed cells. Then we cotransform the two plasmids into DB21 DE3 competent cells. Our device includes an expression vector containing RiAFP cloned downstream of the T7 promoter. DB21 DE3 competent cells carry a chromosomal copy of the phage T7 RNA Polymerase gene, which is controlled by a lac promoter. When inducer (IPTG) is added, T7 RNA Polymerase is expressed and becomes dedicated to transcription of RiAFP.
Plasmid pLG575 includes the coding regions for proteins HlyB and HlyD.
May 20
We are ordering the DB21 DE3 competent cells from NEB very shortly in preparation of cotransformation of these cells with the expression and secretion plasmids.
May 22
We took individual colonies growing on agar supplemented with chloramphenicol and put them in SOB culture broth. SOB is a slightly modified LB broth. The cells, NEB 10 beta cells, transformed with pLG575 plasmid will grow in the broth and provide a source of the plasmid. We also transformed NEB 10 cells with the expression plasmid.
Currently we have liquid cultures of NEB cells with the expression plasmid and liquid cultures of NEB cells with the secretion plasmid. At this point we are poised to do minipreps on both types of cultures.
The minipreps will give us bare plasmids that in turn will be used to transform BL21 DE3 E coli cells. These are chemically competent E. coli cells suitable for transformation and T7 promoter driven expression of RiAFP expression. The two plasmids will be used to cotransform BL21 DE3 cells. Following cotransformation, expression of RiAFP will be induced by IPTG.
May 24
On Tuesday we will do miniprep on the cells containing the expression plasmid. At that point we will be in a position to cotransform the BL21 DE3 cells that constitute the PlantiFreeze device. Competent BL21 DE3 cells will be shipped to the lab on Wednesday on dry ice. We will transform the cells on Wednesday. On Thursday the 16 hour incubation of the transformed cells will be ready. On Thursday, we will take individual colonies from the agar plate and put them into SOC media. On Friday of next week if all goes well we will have an ample supply of the BL21 DE3 cells transformed with both the expression and secretion plasmids. At this point the moment of true has arrived! By introducing IPTG into the culture we should be able to induce the cells to express RiAFP and secrete the recombinant protein into the medium in which the cells are growing. Here is where Steph and the Silver lab come into action. We need to assay the medium and the lyzed cells to confirm the RiAFP was expressed by the E coli and secreted. We will do this using Western blot.
There will be other analyzes that we'll also do:
• Effect of each plasmid and both plasmids on the growth rate of BL21 DE3 cells (by turbidity analysis using spectrophotmometer
• Gel electrophoresis of some of the intermediate constructs
May 24
Rather than using Western blot we are thinking about the use of Invision In-gel stain from Life Technologies. Visualization of the fluorescent bands uses:
UV transilluminator (302 nm) equipped with a camera capable of integration
To view and photograph a gel on the UV transilluminator, use a video camera, CCD (Charged Couple Device) camera, or a cooled CCD camera with ethidium bromide filter or band pass filter encompassing the emission maxima
(590 nm) of the stain.
May 27
DL21 BE3 competent cells from NEB arrive tomorrow. Today we will do miniprep on the cells containing the expression plasmid.
So how 'bout we order:
InVision™ His-Tag In-Gel Staining Kit
InVision™ His-tag In-gel Stain is supplied as a 1X ready-to-use staining reagent (500 mL) [enough to run 20 mini gels]
BenchMark™ His-tagged Protein Standard (125 µL) is also included in the kit
NuPAGE® Novex® 10% Bis-Tris Gels (pre-cast polyacrylamide gels)
May 28
Thanks to Joey and Thomas for doing the miniprep on the expression plasmid. We now have pure DNA from both plasmids and are poised to cotransform BL21 DE3 competent cells. This is the last step in the assembly of the PlantiFreeze device. After that step we do functional assays to confirm that the device works to express and secret RiAFP. As a side venture we'll be exploring the effect of transformation of the BL23 DE 3 cells with each plasmid alone and both together. We'll be constructing growth curves based on optical density of the culture media.
We have ordered from LifeTechnologies Invision in-gel stain, a latter, and precast gels for confirming the expression and secretion of the protein. These experiments will be done with Steph Hayes at Pam Silver's lab at Harvard Medical School.
Keep your fingers crossed that everything goes right and we'll be in the home stretch. Still need to do data analysis, the Wiki, our poster and the presentation so we need everybody at battle stations a while longer.
Finally, we need to do more minipreps more growing up liquid cultures, more transformations so please avail yourself of the opportunity to get your hands wet in the lab. Its fun, its instructional, and you help is really neede
May 29
Lab today 1 to 5:
• cotransformation of Bl21 De 3 cells
• miniprep of plasmids
• making agar plates
• making liquid cultures
• planning measurement of oD of culture for growth curves
May 31
Yesterday was a banner day for lab work. Thanks to Joey, Micah, Aiden, and Thomas. We did a miniprep of the NEB 10 cells transformed with the secretion plasmid pLG575. Did the milestone cotransformation of BL21 DE3 cells with the expression and secretion plasmids. On Saturday after 16 hours incubation at 37 degrees C we should know if it worked. This time we ran a positive control of pUC16 DNA so if something goes wrong will have the control to help us unravel the problem.
If the cotransformation works our next step is to grow up the cotransformed cells in LB broth and then induce them with IPTG to express and secrete RiAFP.
Remember we plan on determining the influence on, the other and both plasmids on the growth of BL21 DE3 cells using the optical density of the liquid culture on the spectrophotometer. If time before the close of the Wiki permits we might also do digestions of the plasmids and look at their electrophoresis patterns.
No growth on either the plates with contransformation or the positive control. It is obviously useful to have a positive control because now we can conclude that we are doing something wrong with the transformation procedure or the competent cells are no longer competent. In fact, in our hands, tansformation has been very unreliable. But this is the first time we ran a positive control.
June 2
The NanoDrop is a specialized spectrophotometer that measures the amount of DNA in a sample as small as 1 microliter.
We are having trouble with the cotransformation and we are worried that the plasmid DNA we are getting from the minipreps was too little. As a benchmark, it is good to have more than 50 nanograms/microliter of plasmid DNA.
We have two plasmids: pLG575 and the expression plasmid. We have 5 nanograms/microliter of this plasmid. Obviously very low. The other plasmid is the expression plasmid. We have 47 nanograms/microliter of this plasmid. Good.
We can adjust the miniprep to increase the concentration of pLG575 DNA,
The puzzle is that we should have gotten transformation of the expression plasmid. So we still have to look at the transformation procedure.
June 6
This has been a frustrating and puzzling week. The cotransformation performed on Sunday failed, leading us to wonder if we got plasmid DNA from the minipreps of the expression and secretion plasmids. Steph generously allowed us to use the Nanodrop analyzer in her lab. We found that the DNA concentration was low but acceptable for pLG575 and in a good range for the expression plasmid. We made some alterations to the miniprep procedure. We repeated the transformations in a wide ranging way and were amazed that the only plate with growth was the one with agar only. This observation directed us to think about a problem with the antibiotics so we made new stock solutions and were especially careful about antibiotic concentrations in the most recent transformations. On Thursday, Joey, Micah, and Thomas were in the lab. We used BL21 DE competent cells and did the following:
• cotransformation with the original pLG575 plasmid from Utah State and the expression plasmid
• transformation of pUC19 positive control plasmid
• transformation with pLG575 plasmid obtained by miniprep using modifications to increase concentration
• transformation of the pPURP plasmid from the BioBuilder lab exercise
All in all, maddeningly frustrating, but we will persevere.
June 7
Puzzling, frustrating, and humiliating! We used commercial BL21 DE3 competent cells in the transformation reactions. pLG575 (orig) refers to the plasmids shipped to use by Utah State; pLG575 (miniprep) refers to the plasmid DNA we got by miniprep of the transformed NEB 10 beta cells transformed with the pLG575 plasmids.
pPURP is the plasmid from BioBuilders and pUC19 is the positive control plasmid sent with the competent cells by NEB.


We mixed new stock solution of ampicillin and chloramphenicol and carefully checked the concentrations in the LB agar, so I am really bewildered by the discrepancies between the expected and observed results.
June 10
Thank you Aiden, Noa, Ben, and Joey for working in Steph's lab at Harvard Medical School. In an attempt at troubleshooting our problem with transformation we did transformations with several different strains of competent cells. The plasmids we used included: pLG575, the expression plasmid, pUC19.
June 11
Yesterday, we used five different strains of competent E coli and five separate plasmids for transformation of the strains of competent cells. Here are the results of the growth of the plates with the appropriate antibiotics:
Results from Steph’s Lab

Disregarding one or two anomalies, the analysis indicates two things:
The BL21 DE3 competent cells don't appear to be competent. Keeping them on dry ice may not have been able to maintain a stable -80 degree centigrade temp allowing them to become incapable of transformation (see column pUC19). Please note that the prior batch of competent cells from NEB (NEB 10 beta cells) were transformed by the pLG575 plasmids we recieved from Utah State and the plasmid we constructed from the gBlocks form IDT. I did not observe thawing of the BL23 DE3 cells. The plasmid DNA we obtained by miniprep were not able to transform any competent cells (see last three columns). The cause of the failure of the miniprep remains uncertain.
June 12
Hi, all. Let’s summarize where we are at the moment.
We have plasmids with the expression cassette (amp resistant) and the secretion cassette (chloramphenicol resistant), respectively in transformed NEB 10 beta cells. This is where we reached a stone wall. Using minipreps, we were unable to cotransform BL21 DE3 cells and induce those cells with IPTG to express and secrete RiAFP. We have an in-gel stain that will enable us to detect RiAFP in cytoplasm and culture media standing by and ready to use (takes about 3 hours to do).
Under the guidance of Steph, team members performed transformations using several E coli strains and different plasmids (the results summarized in an email message sent to all yesterday). The analysis of those results suggest that the commercial competent cells from NEB (BL21 D3) were degrade by unstable maintenance of -80 C temp and that our miniprep procedure was flawed.
It is easy to overcome the competent cell flaw. Not so easy is pin pointing what's wrong with the miniprep. Since we have amp and chloramphenicol resistant NEB 10 cells in the Bio 3 frig, we can analysis the plasmids in those cells and figure out a strategy to go forward. We have in the frig in Bio 3 agar plates and LB broth cultures of the NEB 10 beta cells which are amp resistant cells andchloramphenicol resistant cells meaning they were transformed by the expression and secretion plasmids, respectively. I will bring them to the AlbertsLab tomorrow for analysis confirming the presence of the appropriate plasmids in the cells.
Needless to say, the clock is ticking and it will be a challenge to finish all the lab work. The Wiki closes on 20 June, but the lab work can go on beyond that date and any new data can be incorporated in the Poster and the PowerPoint slide deck. Even then time may run out despite the best efforts.
June 13
Noa's review of the Parts synthesized by IDT prompted me to do a deep dive into the parts sequence synthesized by IDT. Oh boy, did we mess up and compounding that problem we didn't check the parts sequence when they came back from IDT. But since they have a 3-5 business day turn around we can recover but we want to order just one DNA fragment which will be inserted into a linearized plasmid with an antibiotic resistance different from the secretion plasmid. That plasmid will then be cotransformed with our secretion plasmid into BL21 DE3 competent cells. Uncovering the incorrect sequence in the expression plasmid solves our current problem of not being able to cotransform in that we'll have a brand new plasmid and we will use new competent cells.
Here is new revised approach:
we need a plasmid which contains these components and BioBrick standard prefix and suffix (not the RFC 23):
• T7 promoter
• RBS
• RiAFP coding sequence
• HlyA coding sequence
• 6x His marker
• double terminator
We insert (using digestion and ligation) the above fragment into a linearized plasmid with a resistance marker other than chloramphenicol (the secretion plasmid is chloramphenicol resistant). We then use Invision in gel stain to detect presence of RiAFP in cytoplasm and media.
We'll be working up to the last minute: here's the timeline:
• Receive new part from IDT: 19 June
• Insert new part into linearized plasmid: 19 June
• Transform competent cells to grow up stock of plasmid: 20 June
• Grow up liquid culture of transformant: 21 June
• Miniprep of plasmid DNA: 22 June
• Transform BL21 DE3 cells: 22-23 June
• Grow up transformed BL21 DE3 cells and induce expression and secretion of RiAFP: 23-24 June
• Use Invision system to detect RiAFP in fractions: 25 June
• Analyze results: 25-26 June
June 14
After the disheartening discovery that Part A had been incorrectly designed, we decided to engineer a single DNA fragment to serve as the expression genetic circuit in thePlantiFreeze Device. The circuit would comprise the following components:
BioBrick Standard Protein Coding Region Prefix /T7 Promoter /RBS /RiAFP/HlyA /Double terminator /BioBrick Standard Protein Coding Region Suffix
The DNA sequences for the components could be found in the Registry. So it was not difficult to design the part. It was 864 base pairs in length. It is fast and simple to order a 864 base pair gBlock fragment form IDT (double-stranded, sequence-verified genomic blocks that ship in only a few working days for affordable and easy gene construction).
The first step in ordering a gBlock is automated review by screening tools and expert review by IDT scientists of entered sequences for characteristics that may interfere with synthesis. This review found that there were sequence repetitions longer than 8 base pairs that comprise a combined 91% of the sequences within a window of 70 base pairs beginning at base pair 235. This structure would interfere with synthesis. So the IDT gene product specialist optimized the sequence for expression in E coli and compatible with the gBlock synthesis process. Optimization theoretically just changes codon usage without changing the proteins coded. Importantly, the EcoRI and PstI sites on the 5’ and 3’ ends of this gene were maintained compatible with BioBrick standard 3A assembly. However, Noa at Steph's lab ran the optimized new sequence thru the seqbuilder program and identified that the optimization process put in another Pst1 restriction site at position 637 in addition to the appropriate one at position 863 (the one compatible with the BioBrick standard). Having the secondPstI restriction site breaks the BioBrick standard and makes correct assembly impossible. Following the finding of this glitch, the fragment was reoptimizedremoving the PstI site at 836. The part was order for synthesis and delivery as soon as possible.
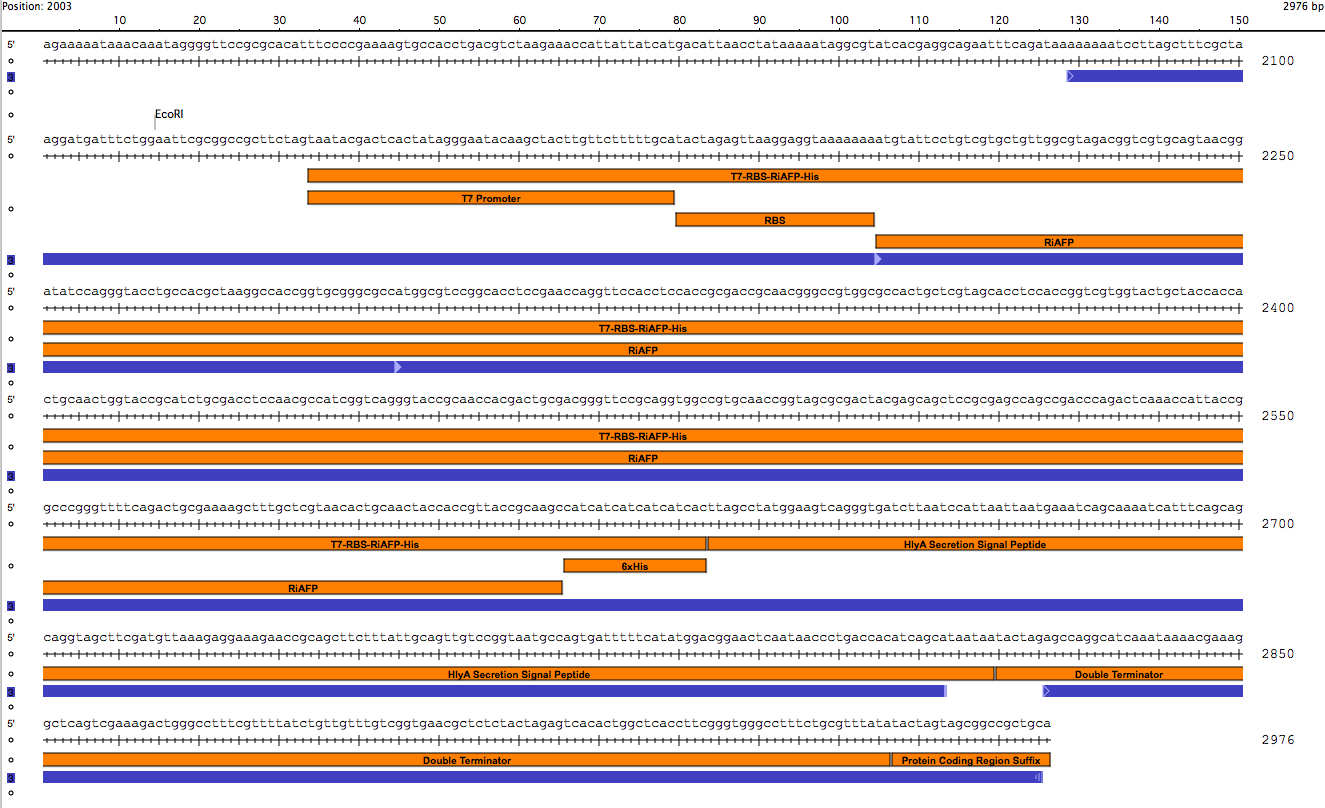
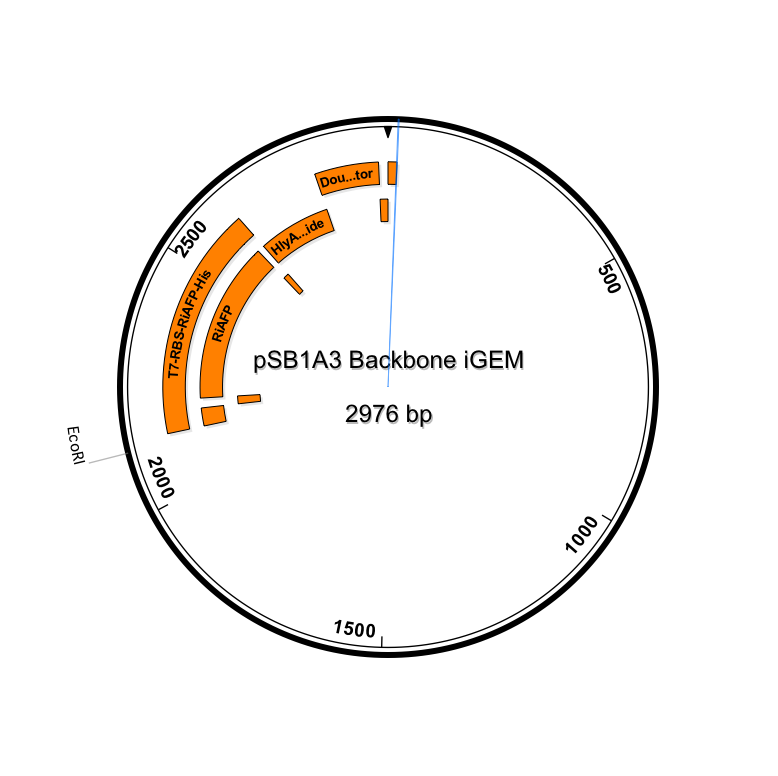
New sequence outlined in SeqBuilder (click to enlarge)
This new part just ordered from IDT would be inserted into a linearized plasmid backbone with antibiotic resistance different from the plasmid controlling secretion (pLG575). We decided to use again pSB1A3. This linearized plasmid is supplied by iGEM. We had exhausted our previous supply so ordered a new one.
The plan is to insert the new part into pSB1A3 and again do cotransformation of BL21 DE3 cells with the two plasmids. Then we would induce thecotransformants with IPTG to express and secrete RiAFP. Finally, using theInvision In-gel Staining Kit we would analyze the media and BL21 DE3 cell lyzatefor RiAFP. This would mean we work up to the very last minute before the Jamboree but we are determined to successfully complete the project.
One last problem is a reliable source of pLG575. Because our mini-centrifuge has a max speed of 4000 rpm (as compared to the recommended of 12000 rpm) our yield of pLG575 from Qiagen miniprep is abysmal (on Nanodrop: 5 ug/uL) The pLG575 generously sent to us by our advisor at Utah State, Asif Rahman, on Nanodrop showed 260 ug/ul. I'll ask Asif to send us another tube of pLG575 from his lab. Also, we will do the miniprep with Steph using a centrifuge of appropriate speed.
So that's our plan! The team is up to the challenge. In the lull while we wait for the new part, we will work on the Wiki due 20 June. and work on the Poster and our 20 minute Presentation.
 "
"