Project.html
From 2014hs.igem.org
AustinWiltse (Talk | contribs) m |
|||
| Line 1: | Line 1: | ||
| - | <html | + | <!DOCTYPE html> |
| + | <html> | ||
| + | <head> | ||
| + | <meta content="text/html; charset=utf-8" http-equiv="content-type"> | ||
| + | <title>GenetiX - The Project</title> | ||
| + | <link href="http://fonts.googleapis.com/css?family=Source+Sans+Pro:300,300italic,600%7CSource+Code+Pro" | ||
| - | + | rel="stylesheet"> | |
| - | < | + | <!--[if lte IE 8]><script src="html5shiv.js" type="text/javascript"></script><![endif]--> |
| - | < | + | <script src="http://code.jquery.com/jquery-1.10.1.min.js"></script> |
| - | < | + | <script src="js/jquery.dropotron.min.js"></script> |
| - | + | <script src="skel.min.js"> | |
| - | + | { | |
| - | + | prefix: 'css/style', | |
| - | + | preloadStyleSheets: true, | |
| - | + | resetCSS: true, | |
| - | + | boxModel: 'border', | |
| - | + | grid: { gutters: 30 }, | |
| - | + | breakpoints: { | |
| - | + | wide: { range: '1200-', containers: 1140, grid: { gutters: 50 } }, | |
| - | + | narrow: { range: '481-1199', containers: 960 }, | |
| - | + | mobile: { range: '-480', containers: 'fluid', lockViewport: true, grid: { collapse: true } } | |
| - | + | } | |
| - | + | } | |
| - | + | </script> | |
| - | + | <script> | |
| - | + | $(function() { | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | } | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | } | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | } | + | |
| - | + | ||
| - | + | ||
| - | } | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | } | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | } | + | |
| - | </ | + | |
| - | < | + | |
| - | + | ||
| - | + | ||
| - | < | + | // Note: make sure you call dropotron on the top level <ul> |
| + | $('#main-nav > ul').dropotron({ | ||
| + | offsetY: -10 // Nudge up submenus by 10px to account for padding | ||
| + | }); | ||
| - | < | + | }); |
| - | + | </script> | |
| + | <script> | ||
| + | // DOM ready | ||
| + | $(function() { | ||
| + | |||
| + | // Create the dropdown base | ||
| + | $("<select />").appendTo("nav"); | ||
| + | |||
| + | // Create default option "Go to..." | ||
| + | $("<option />", { | ||
| + | "selected": "selected", | ||
| + | "value" : "", | ||
| + | "text" : "Menu" | ||
| + | }).appendTo("nav select"); | ||
| + | |||
| + | // Populate dropdown with menu items | ||
| + | $("nav a").each(function() { | ||
| + | var el = $(this); | ||
| + | $("<option />", { | ||
| + | "value" : el.attr("href"), | ||
| + | "text" : el.text() | ||
| + | }).appendTo("nav select"); | ||
| + | }); | ||
| + | |||
| + | // To make dropdown actually work | ||
| + | // To make more unobtrusive: http://css-tricks.com/4064-unobtrusive-page-changer/ | ||
| + | $("nav select").change(function() { | ||
| + | window.location = $(this).find("option:selected").val(); | ||
| + | }); | ||
| + | |||
| + | }); | ||
| + | </script> | ||
| + | </head> | ||
| + | <body> | ||
| + | <div id="header_container"> | ||
| + | <div class="container"> <!-- Header --> | ||
| + | <div id="header" class="row"> | ||
| + | <div class="4u"> | ||
| + | <h1><img src="https://static.igem.org/mediawiki/2014hs/c/c9/Logo.png" alt="" usemap="#Map" height="101" width="301"> | ||
| + | <map name="Map"> | ||
| + | <area shape="rect" coords="-1,19,68,81" href="https://2014hs.igem.org/Main_Page" | ||
| - | + | target="_blank" alt="logo"> | |
| - | < | + | <area shape="rect" coords="68,3,300,103" href="index.html"> |
| - | < | + | </map> |
| - | + | </h1> | |
| - | < | + | </div> |
| - | < | + | <nav id="main-nav" class="8u"> |
| - | < | + | <ul> |
| + | <li><a href="index.html">Home</a></li> | ||
| + | <li><a class="active" href="project.html">The Project</a> | ||
| + | <ul> | ||
| + | <li><a href="project_biosensor.html">Biosensor</a></li> | ||
| + | <li><a href="project_eutrophication.html">Eutrophication</a></li> | ||
| + | <li><a href="project_localization.html">Localization</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="results.html">Results</a> </li> | ||
| + | <li><a href="#">Human Practices</a> | ||
| + | <ul> | ||
| + | <li><a href="human_practices_travesuras2014.html">Travesuras | ||
| + | 2014</a></li> | ||
| + | <li><a href="human_practices_travesuras2013.html">Travesuras | ||
| + | 2013</a></li> | ||
| + | <li><a href="human_practices_conference.html">Conference</a><a | ||
| - | </ | + | href="human_practices_travesuras2014.html"></a></li> |
| - | < | + | </ul> |
| - | < | + | </li> |
| - | < | + | <li> <a href="#">Data</a> |
| - | + | <ul> | |
| - | <p | + | <li><a href="data_safety.html">Safety</a></li> |
| - | + | <li><a href="data_notebook.html">Notebook</a></li> | |
| - | + | <li> <a href="data_biobricks.html">Biobricks</a></li> | |
| - | <p | + | </ul> |
| - | + | </li> | |
| + | <li><a href="#">The Team</a> | ||
| + | <ul> | ||
| + | <li><a href="team_members.html">Members</a></li> | ||
| + | <li><a href="team_sponsors.html">Sponsors</a> </li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | </ul> | ||
| + | </nav> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div id="site_content"> | ||
| + | <div class="container"> | ||
| + | <!-- Banner --> | ||
| + | <div id="banner"> <a href="#"><img src="https://static.igem.org/mediawiki/2014hs/e/ea/Project.jpg" alt="banner image"></a> | ||
| + | </div> | ||
| + | <!-- Features --> | ||
| + | <div class="row"> | ||
| + | <section class="8u"> | ||
| + | <h1>The Project</h1> | ||
| + | <p>The main idea of our project is to achieve the detection of | ||
| + | anoxia and Iron concentrations in water systems. What we propose | ||
| + | is to construct an easy way of monitoring the levels of O2 and Fe | ||
| + | in the lake by using biosensors. By using modified bacteria <em>E. | ||
| + | coli</em> for this, we will try to find a cheaper, easier, | ||
| + | and faster way to detect the problem of anoxia and iron | ||
| + | concentration in some aquifers of Mexico City. We will have to | ||
| + | analyze samples of water at different depths to know where the | ||
| + | problem is worse and what probable native species could be more | ||
| + | affected.</p> | ||
| + | <p><br> | ||
| + | Our purpose is the identification of adequate dissolved oxygen | ||
| + | levels for a stable support of life and the identification of iron | ||
| + | concentration below threatening levels in order to know if the | ||
| + | lake has the physical and chemical properties to support wild-life | ||
| + | naturally. With the use of biosensors, specialized for detecting | ||
| + | concentrations of oxygen above a 2% dissolution, we used modified | ||
| + | E. coli with an oxygen promoter (BBa_K258005) that will | ||
| + | detect the low concentrations and a reporter of (GFP or RFP) that | ||
| + | will indicate the activation of our promoter (<strong>Figure 1</strong>). | ||
| + | The Iron promoter reacts inside an environment with a | ||
| + | concentration of iron ranging from 1 ppm and on (A. | ||
| + | Quintero,2007). The acceptable levels of iron in drinkable water | ||
| + | are lower than 0.5 ppm (WHO, 1996).</p> | ||
| + | <p align="center"><img src="https://static.igem.org/mediawiki/2014hs/0/07/Project_figure1.png" alt="" height="326" | ||
| - | + | width="300"></p> | |
| - | + | <p> To achieve the objective using <em>E. Coli</em> we will | |
| - | <p | + | construct different types of modified plasmids for our bacteria to |
| - | + | express the biosensors based on iGem biobricks. The idea is to use | |
| - | + | sensitive promoters: one for oxygen, and another one for iron; | |
| - | + | those promoters will lead to an expression of GFP or mRFP. This | |
| - | + | will provide a visual signal to indicate the presence or absence | |
| - | + | of these elements. Biobrick parts BBa_K258005 (O₂ prom), | |
| - | + | BBa_I765000 (Fe prom), BBa_E1010 (GFP) and BBa_J04650 (mRFP) were | |
| - | + | selected for the construction of the biosensors (<strong>Figure 2</strong>). | |
| - | + | These were transformed into E. <em>coli</em> strands DH5-a, TOP | |
| - | + | 10, and NEB 10-b for storage and subsequent plasmid growth and | |
| - | + | isolation using a Zymo Research® DNA extraction kit.</p> | |
| - | + | <p align="center"><img src="https://static.igem.org/mediawiki/2014hs/2/22/Project_figure2.png" alt="" height="124" | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <p | + | |
| - | + | ||
| - | <img | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | </ | + | width="350"></p> |
| + | <p> Once we measure and identify critical regions of the Lake, our | ||
| + | report could go directly to the Citizen Council for its | ||
| + | consideration. The competent authorities should be able to propose | ||
| + | concrete legal initiatives to reduce pollution in the identified | ||
| + | areas and start remediation campaigns that re-establish the local | ||
| + | aquatic environment to a stable, liveable, friendly ecosystem for | ||
| + | the inhabitant species. </p> | ||
| + | <h1> </h1> | ||
| + | </section> | ||
| + | <section class="4u"> | ||
| + | <div id="sidebar"> | ||
| + | <section class="12u"> | ||
| + | <h1 class="12u italic">References</h1> | ||
| + | <ul type="disc"> | ||
| + | <li>Chaparro-Herrera, D., Nandini, S., & Sarma, S. (2013). | ||
| + | Effect of water quality on the feeding ecology of axolotl | ||
| + | Ambystoma mexicanum. Journal Of Limnology, 72(3), e46. <a href="http://dx.doi.org/10.4081/jlimnol.2013.e46">http://goo.gl/UzcR6F</a></li> | ||
| + | <li>Khosla, C. & Bailey, J.E (1989) Characterization of | ||
| + | the Oxygen-Dependent Promoter of the Vitreoscilla Hemoglobin | ||
| + | Gene in Escherichia coli. Journal of Bacteriology volume 171 | ||
| + | pp.5995-6004 </li> | ||
| + | <li>Geckil H*, Stark BC, and Webster DA(2001)Cell growth and | ||
| + | oxygen uptake of Escherichia coli and Pseudomonas aeruginosa | ||
| + | are differently affected by the genetically engineered | ||
| + | Vitreoscilla hemoglobin gene.Journal of Biotechnology 2001; | ||
| + | 85(1): 57-66.</li> | ||
| + | <li>A. Quintero (2007), A microbial biosensor device for iron | ||
| + | detection under UV irradiation. Retrieved: 05 of June of | ||
| + | 2014 http://goo.gl/sbVxL1</li> | ||
| + | <li>World Health Organization (1996), Iron in Drinking-Water | ||
| + | Retrieved: 26 of May 2014 http://goo.gl/39EBe4</li> | ||
| + | <li>Alejandro Molina (2010), El ajolote de Xochimilco | ||
| + | http://goo.gl/fEF4MA</li> | ||
| + | <li>Raul Arcos Ramos (2004), Evaluacion de componente organico | ||
| + | como un factor indicativo del estado trofico del lago | ||
| + | Xochimilco <a href="http://www.uaemex.mx/Red_Ambientales/docs/congresos/MORELOS/Extenso/CA/EO/CAO-39.pdf">http://goo.gl/ERp1Nw</a></li> | ||
| + | <li>Davis, John C. "Journal of the Fisheries Research Board of | ||
| + | Canada." <em>Minimal Dissolved Oxygen Requirements of | ||
| + | Aquatic Life with Emphasis on Canadian Species: A Review -</em>. | ||
| + | Journal of the Fisheries Research Board of Canada, 14 Apr. | ||
| + | 2011. Web. 27 May 2014. http://goo.gl/BmvjQG. </li> | ||
| + | </ul> | ||
| + | <p class="12u italic"> </p> | ||
| + | </section> | ||
| + | <section class="12u"> </section> | ||
| + | </div> | ||
| + | </section> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div id="footer_container"> | ||
| + | <div class="container"> | ||
| + | <!-- Footer --> | ||
| + | <footer> | ||
| + | <p><a href="https://twitter.com/GenetiXCCM" target="_blank"><img src="https://static.igem.org/mediawiki/2014hs/f/f7/Twitter.png" | ||
| + | alt="twitter"></a> <a href="https://www.facebook.com/pages/Genetix/231542983696246" | ||
| + | |||
| + | target="_blank"><img src="https://static.igem.org/mediawiki/2014hs/5/55/Facebook.png" alt="facebook"></a> <a | ||
| + | |||
| + | href="mailto:genetixccm@gmail.com?Subject=Hi%20GenetiX" target="_top"><img | ||
| + | |||
| + | src="https://static.igem.org/mediawiki/2014hs/0/09/Rss.png" alt="rss"></a></p> | ||
| + | <p><a href="http://www.itesm.mx/wps/wcm/connect/Campus/CCM/Ciudad+de+Mexico/" | ||
| + | |||
| + | target="_blank"><img src="https://static.igem.org/mediawiki/2014hs/3/3a/Itesm_ccm.png" alt="" height="64" | ||
| + | |||
| + | width="176"></a><a href="http://www.hidrosina.com.mx/new/" target="_blank"><img | ||
| + | |||
| + | src="https://static.igem.org/mediawiki/2014hs/5/5d/Hidrosina.png" alt="" height="32" width="110"></a></p> | ||
| + | <p>Copyright © GenetiX | <a href="http://skeljs.org/">skel.js</a> | <a | ||
| + | |||
| + | href="http://www.css3templates.co.uk">design from | ||
| + | css3templates.co.uk</a></p> | ||
| + | </footer> | ||
| + | </div> | ||
| + | </div> | ||
| + | </body> | ||
</html> | </html> | ||
Revision as of 02:34, 21 June 2014
<!DOCTYPE html>
The Project
The main idea of our project is to achieve the detection of anoxia and Iron concentrations in water systems. What we propose is to construct an easy way of monitoring the levels of O2 and Fe in the lake by using biosensors. By using modified bacteria E. coli for this, we will try to find a cheaper, easier, and faster way to detect the problem of anoxia and iron concentration in some aquifers of Mexico City. We will have to analyze samples of water at different depths to know where the problem is worse and what probable native species could be more affected.
Our purpose is the identification of adequate dissolved oxygen
levels for a stable support of life and the identification of iron
concentration below threatening levels in order to know if the
lake has the physical and chemical properties to support wild-life
naturally. With the use of biosensors, specialized for detecting
concentrations of oxygen above a 2% dissolution, we used modified
E. coli with an oxygen promoter (BBa_K258005) that will
detect the low concentrations and a reporter of (GFP or RFP) that
will indicate the activation of our promoter (Figure 1).
The Iron promoter reacts inside an environment with a
concentration of iron ranging from 1 ppm and on (A.
Quintero,2007). The acceptable levels of iron in drinkable water
are lower than 0.5 ppm (WHO, 1996).

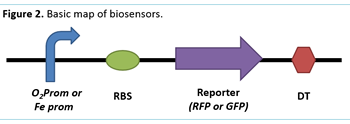
To achieve the objective using E. Coli we will construct different types of modified plasmids for our bacteria to express the biosensors based on iGem biobricks. The idea is to use sensitive promoters: one for oxygen, and another one for iron; those promoters will lead to an expression of GFP or mRFP. This will provide a visual signal to indicate the presence or absence of these elements. Biobrick parts BBa_K258005 (O₂ prom), BBa_I765000 (Fe prom), BBa_E1010 (GFP) and BBa_J04650 (mRFP) were selected for the construction of the biosensors (Figure 2). These were transformed into E. coli strands DH5-a, TOP 10, and NEB 10-b for storage and subsequent plasmid growth and isolation using a Zymo Research® DNA extraction kit.
Once we measure and identify critical regions of the Lake, our report could go directly to the Citizen Council for its consideration. The competent authorities should be able to propose concrete legal initiatives to reduce pollution in the identified areas and start remediation campaigns that re-establish the local aquatic environment to a stable, liveable, friendly ecosystem for the inhabitant species.
 "
"

