
11/13 Our first full-length meeting of the year.
We spent much of the time discussing the logistical (financial, organizational, etc.) details of the iGEM Competition, before splitting off to confer with our small, self-assigned subcommittees: Design, Communications, Fundraising, and Project Management.
11/18
Our lecture half of the class consisted mostly of a detailed overview of synthetic biology, its engineering aspects, and our place in the process.
During the lab half, we learned how to make gels for electrophoresis.
11/20
We spent the first part of class tossing around ideas for a team name; when it came down to either sticking with the school’s mascot (the gryphon) or choosing something more original, a narrow majority vote decided on the latter. Thus, we are now officially the CSW ProTeens.
We then learned more about genetic devices, and specifically their promoters.
11/25
Today, Joey was in sole charge of the class. His lecture covered plasmid backbones in detail. We also watched this CrashCourse video on transcription and translation.
For the lab, we learned (or reviewed) how to make agarose gels, use parafilm, and operate a centrifuge.
12/4
Today’s lecture was on translational control, and the workings of DNA polymerase.
For the lab, we began to work on making our own ampicillin plates from scratch.
12/9
This class was dedicated to finishing our ampicillin plates. We also broke in the new autoclave.
12/11
The team split into smaller groups, and each performed a transformation of pPRL, a purple pigment producing plasmid, into E. coli.
12/16
For the lecture portion of class, we learned about “RNA thermometers,” and then discussed logic gates and used them to analyze some devices.
We then viewed the (successful!) results of our pPRL transformation.
12/18
Joey gave a lecture on 3A assembly.
Our lab was to transform pGRN, a green pigment producing plasmid, into E. coli.
1/6
Happy New Year! We dedicated today to preparing gels for our next lab.
1/8
We began a restriction digest lab, but due to technical difficulties had to postpone until the next class. We then split into committees for a regular update.
1/13
We did some more practice with electrophoresis using lambda DNA.
1/15
We dedicated the whole meeting to practicing with a restriction digest.
1/22
In pairs and singles, we researched and presented the team’s top ten project ideas and began to narrow down that list.
1/27
After the previous week’s presentations, we worked to more fully develop the most feasible ideas and draft circuits for them.
1/29
We gave our final presentations on the most feasible project ideas, which we had been working on for weeks.
2/3
The results are in! We, the CSW ProTeens, will pursue plantifreeze for our final project.
"Plantifreeze" is our shorthand name for a bacteria that prevents frost from forming on fruits near the end of the fall growing season—in other words, a plant antifreeze.
4/27
This weekend we made stock solutions of antibiotic solutions, prepared sterile LB agar plates supplemented with ampicillin, chloramphenicol, and the combination. We also prepared SOB broth for growing up transformed bacteria.
Consequently, we are ready now to begin assembly of our device.
4/29
Today we transformed competent NEB-10 Beta Cells with pLG575 (secretion plasmid) to have a large supply of plasmid obtainable by mini-prep.
4/30
We took the gBlocks of parts A and B of the expression plasmid and the linearized plasmid pSB3A1 and digested the prefix and suffix of each with restriction enzymes: EchoRI, SpeI, Xbal, PstI, PslI. These enzymes recognize restriction sites and cut them in ways that allow part A to fuse with part B and for the fusion of A-B to fit into pSB3A1. The next step is to permanently ligate the parts so that we end up with a circular plasmid carrying the fusion part. The ligation will be performed on Thursday morning between 10 am and 1 pm.
5/1
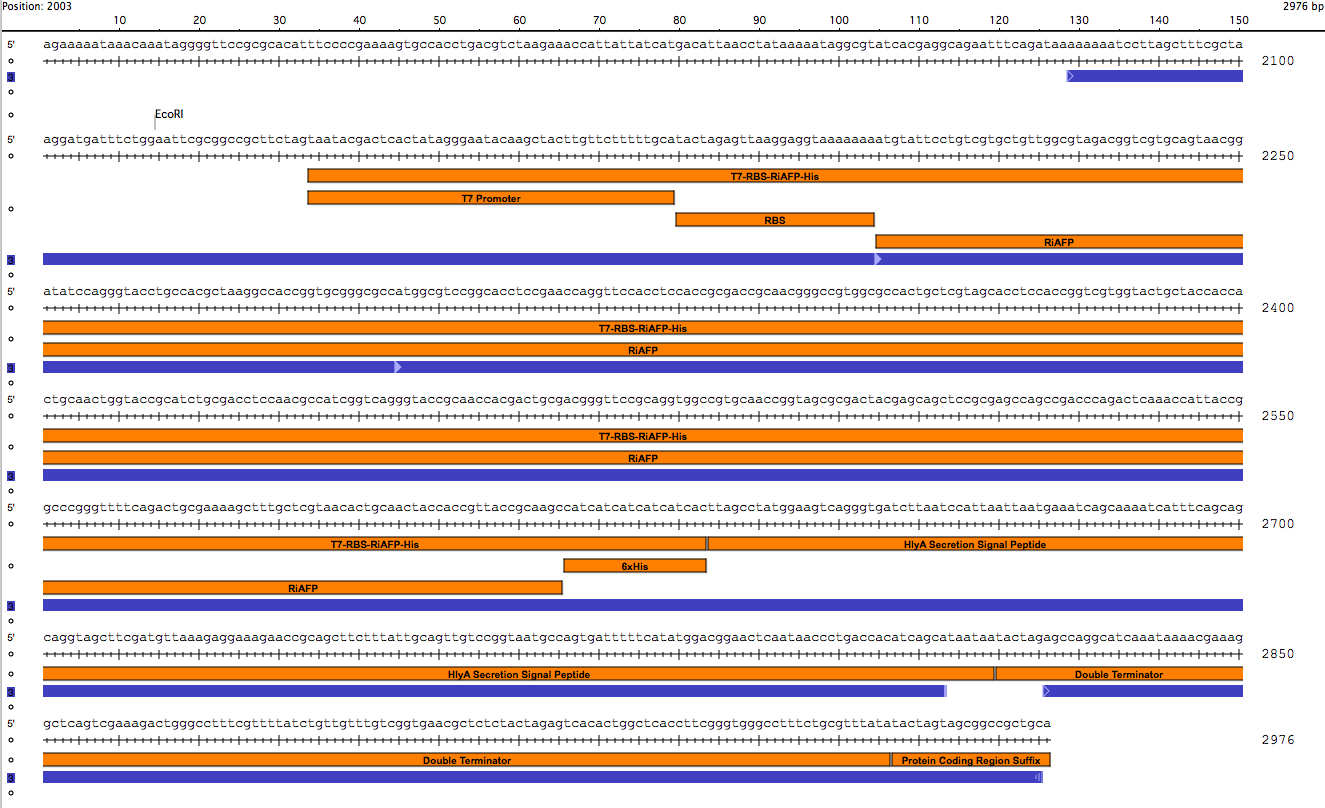
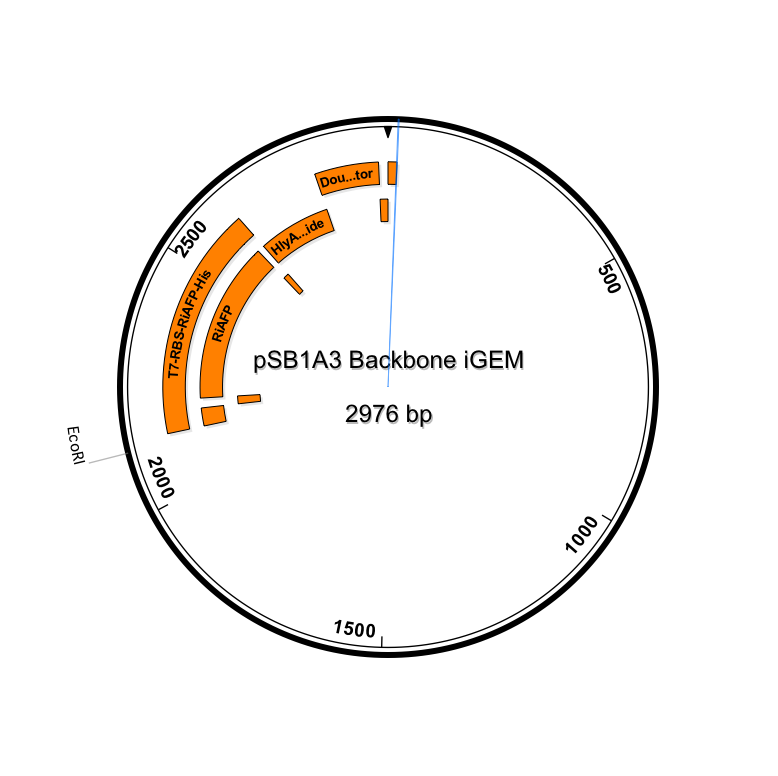
Yesterday we cut Parts A and B and pSB3A1 so that they would would together. Today, we ligated the parts into our expression plasmid; T7 promoter RBS RiAFP 6xHis HlyA double terminator. The next step is to transform NEB-10 Beta competent cells with the plasmid.
5/4
We transformed our expression plasmid into NEB-10 Beta cells.
5/6
The transformation failed. We later discovered that we used 0.2 micrograms of plasmid instead of the suggested 1 microgram while constructing the expression plasmid from parts A and B. This altered the volume of the reaction, and was likely the cause of failure.
5/8
Today we took individual colonies of NEB 10 cells transformed with pLG575 plasmids and put them in LB broth cultures to incubate at 20 degrees C for 24 hours. Growing up these cells will afford us a large supply of the pLG575 plasmids. We ordered more gBlocks from IDT. We will carefully rewrite the protocol for digestion and ligation of the parts that go into the linearized pSB3A1 so transformation can be successful with a future attempt. Also we will run concurrent positive controls in the future to avoid our inability to analyze any future failures.
5/14
After very careful preparation, the parts for the expression plasmid were digested, ligated and the resulting ligation product was used to transform NEB 10 cells. Many things were changed from the unsuccessful transformation from last week. The transformed cells are in the 37 degree C incubator so please everyone hold your breath. We worked on the strain 4-1 and 4-2 E coli that will be transformed with two plasmids to produce green and purple fluorescent cells under UV light (for our Human Outreach Illuminarium stand).
5/15
The transformation of our expression plasmid was successful! We have the expression plasmid in NEB 10 beta cells on agar supplemented with amp. The next step is to grow up colonies from the agar plate in LB broth. This will give us a large supply of plasmids. We will miniprep the liquid culture getting bare plasmids for contranformation of DB21 DE3 competent cells with both plasmids. Then by using Western blot we can confirm presence of RiAFP in supernatant and lysate. In the ligation step of constructing the expression plasmid, we were very careful to use the correct proportion of DNA from the inserts (Parts A and B) and the vector (linearized pSB1A3). We used 50 ng of vector DNA and made use of a calculator on the NEB web site to calculate the amount of insert DNA to use. There should be a 3 to 1 molar excess of insert DNA to vector DNA to ensure that all the linearized vector is fully occupied by insert DNA. The calculation used the nucleotide lengths of the inserts (about 450 bp for each part) and the vector (2155 bp) to make the calculation. Also for the incubation in the ligation step we incorrectly used 22 degrees C instead of room temperature. This was corrected in the latest transformation.
5/16--5/17
After we transformed the NEB 10 beta cells with the newly assembled plasmid (T7 RBS RiAFP 6xHis HlyA term term), we selected a few individual colonies from the agar plate supplemented with amp and put them in 50 mL of LB broth supplemented with amp in sterile 250 ml flasks with a sterile magnetic stirrer and grew up the cells over 16 hours at room temperature with gentle stirring. Our newly acquired rotating shaker has a broken belt so it is on the bench. After the 16 hour incubation, the broth had turned from clear to opaque. We transferred 10 ml of the broth to sterile falcon tubes and stored them at 4 degrees C. We do minipreps on the cells to isloate and purify the plasmids. We also transformed NEB 10 cells wit the pLG575 plasmids and stored them in 10 ml broth at 4 degrees C. So now we have ample supplies of both plasmids. The next step is to miniprep the transformed cells. Then we cotransform the two plasmids into DB21 DE3 competent cells. Our device includes an expression vector containing RiAFP cloned downstream of the T7 promoter. DB21 DE3 competent cells carry a chromosomal copy of the phage T7 RNA Polymerase gene, which is controlled by a lac promoter. When inducer (IPTG) is added, T7 RNA Polymerase is expressed and becomes dedicated to transcription of RiAFP. Plasmid pLG575 includes the coding regions for proteins HlyB and HlyD.
5/21
We took individual colonies growing on agar supplemented with chloramphenicol and put them in SOB culture broth. SOB is a slightly modified LB broth. The cells, NEB 10 beta cells, transformed with pLG575 plasmid will grow in the broth and provide a source of the plasmid. We also transformed NEB 10 cells with the expression plasmid. Currently we have liquid cultures of NEB cells with the expression plasmid and liquid cultures of NEB cells with the secretion plasmid. At this point we are poised to do minipreps on both types of cultures. The minipreps will give us bare plasmids that in turn will be used to transform BL21 DE3 E coli cells. These are chemically competent E. coli cells suitable for transformation and T7 promoter driven expression of RiAFP expression. The two plasmids will be used to cotransform BL21 DE3 cells. Following cotransformation, expression of RiAFP will be induced by IPTG.
5/27
Today we did another miniprep of the expression plasmid to keep our inventory up.
5/28
We now have pure DNA from both plasmids and are poised to cotransform BL21 DE3 competent cells. This is the last step in the assembly of the PlantiFreeze device. After that step we do functional assays to confirm that the device works to express and secret RiAFP. As a side venture we'll be exploring the effect of transformation of the BL23 DE 3 cells with each plasmid alone and both together. We'll be constructing growth curves based on optical density of the culture media. We ordered from LifeTechnologies Invision in-gel stain, a ladder, and precast gels for confirming the expression and secretion of the protein. These experiments will be done at Harvard Medical School with our HMS advisor.
5/30
Today we taught people about our project and synthetic biology at Illuminarium, a school event open to all. Check it out in our Human Outreach section! We did a miniprep of the NEB 10 cells transformed with the secretion plasmid pLG575. We did the cotransformation of BL21 DE3 cells with the expression and secretion plasmids. On Saturday after 16 hours incubation at 37 degrees C we should know if it worked. This time we ran a positive control of pUC 16 DNA so that if something goes wrong we will have the control to help us understand the problem. If the cotransformation works our next step is to grow up the cotransformed cells in LB broth and then induce them with IPTG to express and secrete RiAFP.
6/1
We saw no growth on both the plates with contransformation or the positive control. It is useful to have a positive control because now we can conclude that we are doing something wrong with the transformation procedure. Transformation has been very unreliable for us. This is the first time we ran a positive control. Because the overall time of the procedure is about 3.5 hours we tend to rush and can make mistakes. Below is a new protocol that we will now adopt:
1. Thaw (30 minutes) competent cells on wet ice after removing from -70°C
2. Mix competent cells gently by lightly flicking the tube.
3. Add DNA solution (≤5μl per 50μl cells: for cotransformation add 1μl ) to competent cells and gently swirl tube(s) for a few seconds to mix. If a control is desired, repeat this step with 2μl of the provided pUC19 in a separate tube.
4. Incubate on ice for 30 minutes.
5. Place tube(s) in 42°C water bath for ~30 to 45 seconds without shaking
Note: The heat shock step is calibrated to the size and material of these tubes. If different tubes are used, the heat transfer to the cells may not be optimal.
6. Place tube(s) again on ice for ~5 minutes.
7. Dilute transformation reaction(s) to 1ml by addition of 500μl SOC medium
8. Shake tube(s) ~200 rpm for 2 hours at 37°C.
Note: This outgrowth step allows the bacteria time to generate the antibiotic resistance proteins encoded on the plasmid backbone so that they will be able to grow once plated on the antibiotic containing agar plate. This step is not critical for Ampicillin resistance but is much more important for other antibiotic resistances.
9. Plate by spreading 5-200μl of cell transformation mixture on LB agar plates containing
appropriate antibiotic and incubate overnight at 37°C.
6/2
We are having trouble with the cotransformation and were worried that the plasmid DNA we are getting from the minipreps was too low in quantity. As a benchmark, it is good to have more than 50 nanograms/microliter of plasmid DNA. We have two plasmids: pLG575 and the expression plasmid. We have 5 nanograms/microliter of pLG575. Very low. The other plasmid is the expression plasmid. We have 47 nanograms/microliter of this plasmid. We will adjust the miniprep to increase the concentration of pLG575 DNA. The biggest puzzle is that we should have gotten transformation of the expression plasmid, so we still have to look at the transformation procedure.
6/3
Based on the low concentration in the secretion plasmid DNA, we will decrease the volume of EB buffer we use from 750 micrograms to 250, let the EB buffer sit in the spin column for 5 minutes before centrifuging, and warm the EB buffer. These maneuvers will increase the concentration of DNA when we elute. For today, we used this modified technique in the cotransformation. Importantly, we eliminated the 1 hour incubation in wet ice after the 2 hr 37 C incubation. In addition to cotransformation we transformed cells with each of the plasmids individually. We also made more plates.
6/6
The cotransformation performed on Sunday failed, leading us to wonder if we got plasmid DNA from the minipreps of the expression and secretion plasmids. Steph (our advisor) generously allowed us to use the Nanodrop analyzer in her lab. We found that the DNA concentration was low but acceptable for PLG575 and in a good range for the expression plasmid. We made some alterations to the miniprep procedure. We repeated the transformations in a wide ranging way and were amazed that the only plate with growth was the one with agar only. This observation directed us to think about a problem with the antibiotics so we made new stock solutions and were especially careful about antibiotic concentrations in the most recent transformations. On Thursday we used BL21 DE competent cells and did the following: a cotransformation with the original pLG575 plasmid from Utah State and the expression plasmid, a transformation of pUC19 positive control plasmid, a transformation with pLG575 plasmid obtained by miniprep using modifications to increase concentration, and a transformation of the pPURP plasmid from the BioBuilder lab exercise.
6/10
Our last transformations all failed as well. We went to the Wyss Institute to meet up with Steph, who let us use 3 new types of competent cells. Our analysis of the results from our day at Wyss tell us that the BL21 DE3 competent cells don't appear to be competent. Keeping them on dry ice may not have been able to maintain a stable -80 degree centigrade temp allowing them to become incapable of transformation (see column pUC19). The prior batch of competent cells from NEB (NEB 10 beta cells) were transformed by the pLG575 plasmids we recieved from Utah State and the plasmid we constructed from the gBlocks form IDT. We did not observe thawing of the BL23 DE3 cells. The plasmid DNA we obtained by miniprep were not able to transform any competent cells. The cause of the failure of the miniprep remains uncertain.
6/11
After using SeqBuilder software to analyze our parts from our expression plasmid, we found that a deletion of a base pair between Part A and B lead to a shift of frame and an early termination. We rebuilt our plasmid and will now be synthesizing a new gBlock that includes parts A and B. The open reading frames and restriction sites are still in place.

 "
"

