Team:HUNGENIOUS/Bioinfo
From 2014hs.igem.org
(Difference between revisions)
| (8 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | <style type="text/css"> | ||
<html> | <html> | ||
<head> | <head> | ||
| - | < | + | <map id="logomap" name="logomap"> |
| - | < | + | <area shape="rect" alt="" title="" coords="778,111,961,281" href="https://2014hs.igem.org" target="iGEM" /> |
| - | < | + | </map> |
| - | < | + | |
| - | <style type="text/css"> | + | <script type="text/javascript" src="http://igem2.mabite.info/jscookmenu.js"></script> |
| - | + | <script type="text/javascript" src="http://igem2.mabite.info/ThemeAqua/theme.js"></script> | |
| - | { | + | <script type="text/javascript" src="http://igem.mabite.info/csongor/header.js"></script> |
| - | + | <link rel="stylesheet" href="http://igem.mabite.info/csongor/style.css" type="text/css"/> | |
| - | + | ||
| - | + | <style type="text/css"> | |
| - | + | #content { | |
| - | + | background-color: #01B4D3; | |
| - | } | + | color:#01B4D3; |
| - | </style> | + | scrollbar-face-color:#01B4D3; |
| - | < | + | scrollbar-arrow-color:#01B4D3; |
| - | + | scrollbar-3dlight-color:#ECE9D8; | |
| + | scrollbar-darkshadow-color:#716F64; | ||
| + | scrollbar-highlight-color:#01B4D3; | ||
| + | scrollbar-shadow-color:#ACA899; | ||
| + | scrollbar-track-color:#D4D0C8; | ||
| + | } | ||
| + | |||
| + | a { | ||
| + | color:#7D9EC0; | ||
| + | } | ||
| + | |||
| + | a:visited { | ||
| + | color: #7B7BC0; | ||
| + | } | ||
| + | |||
| + | a:hover { | ||
| + | color:#1AFBCB; | ||
| + | } | ||
| + | </style> | ||
| + | </head> | ||
| + | |||
| + | <body> | ||
| + | |||
| + | <script type="text/javascript" src="http://igem.mabite.info/csongor/menu.js"></script> | ||
| + | |||
{ | { | ||
background-color: #66CCFF; | background-color: #66CCFF; | ||
| Line 30: | Line 55: | ||
</style> | </style> | ||
</head> | </head> | ||
| + | |||
<body> | <body> | ||
| + | <style type="text/css"> | ||
| + | div#container | ||
| + | { | ||
| + | width: 800px; | ||
| + | height: 2525px; | ||
| + | margin-top: 0px; | ||
| + | margin-left: 0px; | ||
| + | text-align: left; | ||
| + | } | ||
| + | </style> | ||
| + | <div id="tartalom"> | ||
| + | <div id="container"> | ||
<div id="container"> | <div id="container"> | ||
<div id="bv_Text1" style="margin:0;padding:0;position:absolute;left:24px;top:25px;width:925px;height:39px;text-align:center;z-index:0;"> | <div id="bv_Text1" style="margin:0;padding:0;position:absolute;left:24px;top:25px;width:925px;height:39px;text-align:center;z-index:0;"> | ||
| - | <font style="font-size:32px" color="#000000" face="Calibri"><b>In silico treasure hunt</b></font></div> | + | <font style="font-size:32px" color="#000000" face="Calibri"><b><br>In silico treasure hunt</b></font></div> |
<div id="bv_Text2" style="margin:0;padding:0;position:absolute;left:57px;top:78px;width:901px;height:138px;text-align:justify;z-index:1;"> | <div id="bv_Text2" style="margin:0;padding:0;position:absolute;left:57px;top:78px;width:901px;height:138px;text-align:justify;z-index:1;"> | ||
<font style="font-size:19px" color="#000000" face="Calibri">Nowadays bioinformatics become more and more important along with the continuously developing bioinformatics databases. We have checked our target gene - what type of information can we find about it.<br> | <font style="font-size:19px" color="#000000" face="Calibri">Nowadays bioinformatics become more and more important along with the continuously developing bioinformatics databases. We have checked our target gene - what type of information can we find about it.<br> | ||
| Line 58: | Line 96: | ||
<div id="bv_Image4" style="margin:0;padding:0;position:absolute;left:17px;top:2251px;width:948px;height:370px;text-align:left;z-index:8;"> | <div id="bv_Image4" style="margin:0;padding:0;position:absolute;left:17px;top:2251px;width:948px;height:370px;text-align:left;z-index:8;"> | ||
<img src="http://igem2.mabite.info/images/bioinf_4.jpg" id="Image4" alt="" align="top" border="0" style="width:948px;height:370px;"></div> | <img src="http://igem2.mabite.info/images/bioinf_4.jpg" id="Image4" alt="" align="top" border="0" style="width:948px;height:370px;"></div> | ||
| - | </div> | + | </div></div></div> |
</body> | </body> | ||
</html> | </html> | ||
Latest revision as of 07:51, 15 July 2014
<style type="text/css">
{ background-color: #66CCFF; background-image: url(images/bioinfo_bkgrnd.png); color: #000000; }In silico treasure hunt
Nowadays bioinformatics become more and more important along with the continuously developing bioinformatics databases. We have checked our target gene - what type of information can we find about it.
Initially we have tried a random search on the phrase "chitinase" at http://www.ebi.ac.uk/. The results are impressive: almost 40 thousands of several nucleotide sequennces and 32.000 protein sequences are in connection with chitinase.
Initially we have tried a random search on the phrase "chitinase" at http://www.ebi.ac.uk/. The results are impressive: almost 40 thousands of several nucleotide sequennces and 32.000 protein sequences are in connection with chitinase.
If we look for Chromobacterium violaceum chitinase we receive four genomic data. If we specify our search for Cv_2935 gene we can find our target gene at http://www.ensembl.org/index.html, where we can find tremendous amount of data on this gene.
Some of the data:
- Our target gene (cv_2935) can be found on the reverse strand of the chromosome of Chromobacterium violaceum between the 3,211,716th and the 3,213,035th bp on the reverse strand.
- The length of the gene is 1320 bp which codes 439 amino acids.
- There are two further code-numbers here: AAQ60603 for the mRNA transcript and the same number for the protein itself.
- We can find also our gene's neighbours as well.
On this page we also find a further link to the sequence of the gene:
- Our target gene (cv_2935) can be found on the reverse strand of the chromosome of Chromobacterium violaceum between the 3,211,716th and the 3,213,035th bp on the reverse strand.
- The length of the gene is 1320 bp which codes 439 amino acids.
- There are two further code-numbers here: AAQ60603 for the mRNA transcript and the same number for the protein itself.
- We can find also our gene's neighbours as well.
On this page we also find a further link to the sequence of the gene:
If we run a BLAST search for nucleotide sequences we will find that there are many organisms with similar genes. If we look the results carefully we find that there are many organisms with similar chtinase genes with similar sequences. It's really interesting for example, that Streptomyces olivaceoviridis has almost the same DNA sequence for the same function. Compared with other data it might has a meaning that the Chromobacterium violaceum chitinase gene was rather conservative sequence during the evolution.



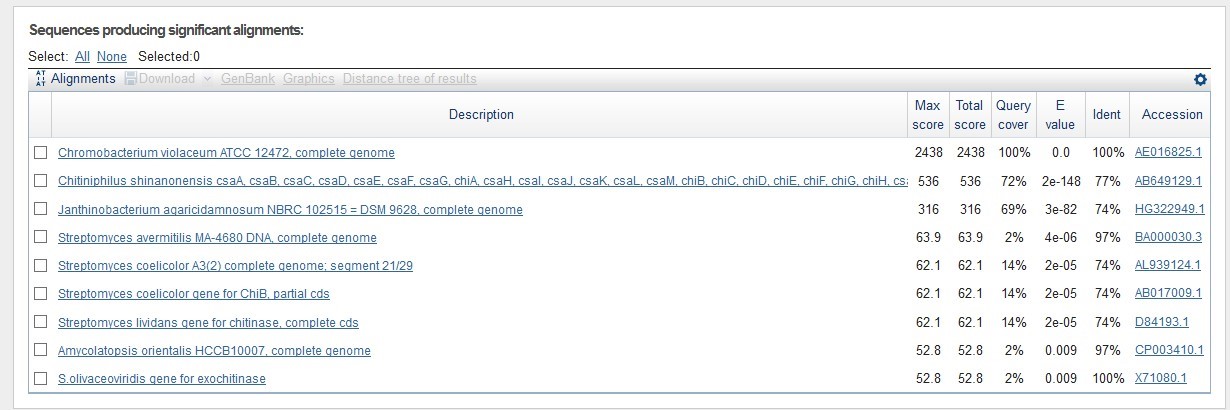
 "
"