Team:CSWProteens/notebook/parts
From 2014hs.igem.org
| (8 intermediate revisions not shown) | |||
| Line 12: | Line 12: | ||
.auto-style1 { | .auto-style1 { | ||
font-family: Arial; | font-family: Arial; | ||
| - | font-size: | + | font-size: 12pt; |
margin-top: 0px; | margin-top: 0px; | ||
margin-bottom: 0px; | margin-bottom: 0px; | ||
margin-right: 0px; | margin-right: 0px; | ||
margin-left: 0px; | margin-left: 0px; | ||
| + | color: #000000 | ||
} | } | ||
| Line 129: | Line 130: | ||
<meta content="revealTrans(Duration=3.0,Transition=2)" http-equiv="Site-Enter" /> | <meta content="revealTrans(Duration=3.0,Transition=2)" http-equiv="Site-Enter" /> | ||
</head> | </head> | ||
| - | <center><a href="https://2014hs.igem.org/Team:CSWProteens/project"><img alt="" class="auto-style11" height="200" src="https://static.igem.org/mediawiki/2014hs/c/cd/Plantifreeze.gif" width="200" /><img src="https://static.igem.org/mediawiki/2014hs/ | + | <center><a href="https://2014hs.igem.org/Team:CSWProteens/project"><img alt="" class="auto-style11" height="200" src="https://static.igem.org/mediawiki/2014hs/c/cd/Plantifreeze.gif" width="200" /><img src="https://static.igem.org/mediawiki/2014hs/7/7e/CSW_Banner.png" width="500"> |
</a> | </a> | ||
| Line 171: | Line 172: | ||
<a href="https://2014hs.igem.org/Team:CSWProteens/humanpractices"><b>Outreach</b></a> | <a href="https://2014hs.igem.org/Team:CSWProteens/humanpractices"><b>Outreach</b></a> | ||
<ul> | <ul> | ||
| - | + | ||
</ul> | </ul> | ||
</li> | </li> | ||
| Line 183: | Line 184: | ||
</ul> | </ul> | ||
<center><p class="auto-style4">P A R T S </center> | <center><p class="auto-style4">P A R T S </center> | ||
| + | <p><p class="auto-style1"> | ||
| + | Part: <a href="http://parts.igem.org/cgi/partsdb/part_info.cgi?part_name=BBa%20K1251000">BBa_K1251000</a> | ||
| - | + | <p><p class="auto-style1"> | |
| - | + | ||
| - | + | ||
• The project undertaken by the Cambridge School of Weston (CSW) 2014 iGEM team builds upon the work of the 2009 Utah State team that developed a protein secretion mechanism and that of the 2011 Yale team which synthesized an “antifreeze” protein (RiAFP) isolated from a cold-tolerant beetle called Rhagium inquisitor. | • The project undertaken by the Cambridge School of Weston (CSW) 2014 iGEM team builds upon the work of the 2009 Utah State team that developed a protein secretion mechanism and that of the 2011 Yale team which synthesized an “antifreeze” protein (RiAFP) isolated from a cold-tolerant beetle called Rhagium inquisitor. | ||
| + | <center> | ||
| - | + | <img src="https://static.igem.org/mediawiki/2014hs/2/2e/Part1.png" width="600"></center> | |
| - | + | <p><p class="auto-style1"> | |
| - | + | ||
• To save time and simplify construction, the part was made by direct synthesis and ordered as gBlocks™ from IDT. gBlocks Gene Fragments are custom, double-stranded, sequence-verified fragments of DNA up to 700 bp. | • To save time and simplify construction, the part was made by direct synthesis and ordered as gBlocks™ from IDT. gBlocks Gene Fragments are custom, double-stranded, sequence-verified fragments of DNA up to 700 bp. | ||
| - | + | <center> | |
| + | <img src="https://static.igem.org/mediawiki/2014hs/8/8a/GBlock_Restriction.png" width="600"></center> | ||
| + | <p><p class="auto-style1"> | ||
| - | + | • We directly synthesized a composite part based on parts from other teams<br> | |
| - | • We directly synthesized a composite part based on parts from other teams | + | • There are no illegal restriction sites (EcoR1, Xba1, Spe1, Pst1) that would interfere with BioBrick standard assembly. The part is flanked by the BioBrick prefix and suffix.<br> |
| - | • There are no illegal restriction sites (EcoR1, Xba1, Spe1, Pst1) that would interfere with BioBrick standard assembly. The part is flanked by the BioBrick prefix and suffix. | + | • The gBlock fragment inserted into a Registry supported plasmid backbone.<p><center> |
| - | • The gBlock fragment inserted into a Registry supported plasmid backbone. | + | <img src="https://static.igem.org/mediawiki/2014hs/5/54/Part_Into_Plasmid.jpg" width="600"></center> |
| - | + | ||
| + | <p><p class="auto-style1"> | ||
| - | • The part controls RiAFP expression which is tagged with a 6x His motif and a HlyA secretion signal. Initiation of transcription of RiAFP/6xHis/HlyA is controlled by a phage promoter, T7. A double terminator stops transcription | + | • The part controls RiAFP expression which is tagged with a 6x His motif and a HlyA secretion signal. Initiation of transcription of RiAFP/6xHis/HlyA is controlled by a phage promoter, T7. A double terminator stops transcription<p><center><img src="https://static.igem.org/mediawiki/2014hs/5/51/New_BioBrick_Part1.jpg" width="600"></center> |
| - | + | ||
| - | + | ||
| - | + | ||
| + | <p><p class="auto-style1"> | ||
| + | <b>Promoter. </b>Our device uses a T7 system containing a lac operator, as did the Yale team, because T7 RNA polymerase is an incredibly fast and powerful enzyme transcribing rapidly and profusely for as long as the T7 RNA polymerase is present. It synthesizes RNA at a rate several times that of E. coli RNA polymerase and it terminates transcription less frequently. Expression can only be achieved in a bacterial strain carrying the gene for the T7 RNA polymerase. The most common cell strain to use with a T7 promoter system is BL21(DE3) (competent cells are chemically competent cells used for high-level protein expression with T7 RNA polymerase-based expression systems). The BL21(DE3) contains the T7 RNA polymerase gene, under the control of the lacUV5 promoter, integrated into the chromosome. IPTG is used to induce the expression of recombinant proteins cloned into vectors downstream of a T7 RNA promoter and transformed into the BL21(DE3) cells. | ||
| + | <p><p class="auto-style1"> | ||
The T7 promoter is a BioBrick part BBa_1712074 | The T7 promoter is a BioBrick part BBa_1712074 | ||
| - | + | <p><p class="auto-style1"><b> | |
| - | C-terminal His-tag. In our protein expression construct, there is an amino acid motif that consists of six histidine (His) residues fused to the C-terminus of RiAFP. Histidine tags are widely used because they are small and rarely interfere with the function, activity, or structure of target proteins. The polyhistidine-tag is to be used to detect the secreted protein via anti-polyhistidine-tag antibodies or alternatively by in-gel staining (SDS-PAGE) with fluorescent probes bearing metal ions. | + | C-terminal His-tag. </b>In our protein expression construct, there is an amino acid motif that consists of six histidine (His) residues fused to the C-terminus of RiAFP. Histidine tags are widely used because they are small and rarely interfere with the function, activity, or structure of target proteins. The polyhistidine-tag is to be used to detect the secreted protein via anti-polyhistidine-tag antibodies or alternatively by in-gel staining (SDS-PAGE) with fluorescent probes bearing metal ions.<p> <p class="auto-style1"> |
| - | + | <b> | |
| - | HlyA-signal peptide. The HlyA is a signal peptide found in the C-terminal signal sequence of alpha-hemolysin (HlyA). It is used to target RiAFP for secretion via the Type I secretion pathway of gram-negative bacteria. Fusion of the HlyA signal peptide to RiAFP results in transport of the protein from the cytoplasm to the extracellular medium in a single step. | + | HlyA-signal peptide. </b>The HlyA is a signal peptide found in the C-terminal signal sequence of alpha-hemolysin (HlyA). It is used to target RiAFP for secretion via the Type I secretion pathway of gram-negative bacteria. Fusion of the HlyA signal peptide to RiAFP results in transport of the protein from the cytoplasm to the extracellular medium in a single step. |
| - | + | <p><p class="auto-style1"> | |
The HlyA signal peptide is BioBrick pat BBa_K208006 | The HlyA signal peptide is BioBrick pat BBa_K208006 | ||
| - | + | <p><b><p class="auto-style1"> | |
| - | Terminator. There are several E. coli transcriptional terminators available via the Registry. The most commonly used type of terminator is a forward terminator. When placed downstream of a genetic part that is transcribed, a forward transcriptional terminator will cause transcription to abort. We use part BBa_B0015, a double terminator (B0010-B0012). BBa_B0015. This terminator has an Average Forward Termination Efficiency of 98.4%. | + | Terminator. </b>There are several E. coli transcriptional terminators available via the Registry. The most commonly used type of terminator is a forward terminator. When placed downstream of a genetic part that is transcribed, a forward transcriptional terminator will cause transcription to abort. We use part BBa_B0015, a double terminator (B0010-B0012). BBa_B0015. This terminator has an Average Forward Termination Efficiency of 98.4%. |
| - | + | <p> | |
| - | + | <p class="auto-style1"> | |
| - | Our PlantiFreeze device is a dual plasmid system in BL21 DE3 E coli. | + | Our PlantiFreeze device is a dual plasmid system in BL21 DE3 E coli.<p><center><img src="https://static.igem.org/mediawiki/2014hs/a/ab/Dual_Plasmid_System.png" width="600"><p><center><a href="https://static.igem.org/mediawiki/2014hs/1/10/IGEM_Seqbuilder_Expression_plasmid_in_backbone.png"><img src="https://static.igem.org/mediawiki/2014hs/1/10/IGEM_Seqbuilder_Expression_plasmid_in_backbone.png" width="450"></a> <a href="https://static.igem.org/mediawiki/2014hs/6/61/Screen_shot_2014-06-13_at_11.04.23_AM.png"> <img src="https://static.igem.org/mediawiki/2014hs/6/61/Screen_shot_2014-06-13_at_11.04.23_AM.png" width="450"></a><br><p class="auto-style1"><b>Sequence outlined in SeqBuilder (click to enlarge). The image on the left is the single-strand DNA sequence of the part. The image on the right is the plasmid map. </b></center> |
Latest revision as of 02:16, 21 June 2014



P A R T S
Part: BBa_K1251000
• The project undertaken by the Cambridge School of Weston (CSW) 2014 iGEM team builds upon the work of the 2009 Utah State team that developed a protein secretion mechanism and that of the 2011 Yale team which synthesized an “antifreeze” protein (RiAFP) isolated from a cold-tolerant beetle called Rhagium inquisitor.

• To save time and simplify construction, the part was made by direct synthesis and ordered as gBlocks™ from IDT. gBlocks Gene Fragments are custom, double-stranded, sequence-verified fragments of DNA up to 700 bp.

• We directly synthesized a composite part based on parts from other teams
• There are no illegal restriction sites (EcoR1, Xba1, Spe1, Pst1) that would interfere with BioBrick standard assembly. The part is flanked by the BioBrick prefix and suffix.
• The gBlock fragment inserted into a Registry supported plasmid backbone.

• The part controls RiAFP expression which is tagged with a 6x His motif and a HlyA secretion signal. Initiation of transcription of RiAFP/6xHis/HlyA is controlled by a phage promoter, T7. A double terminator stops transcription

Promoter. Our device uses a T7 system containing a lac operator, as did the Yale team, because T7 RNA polymerase is an incredibly fast and powerful enzyme transcribing rapidly and profusely for as long as the T7 RNA polymerase is present. It synthesizes RNA at a rate several times that of E. coli RNA polymerase and it terminates transcription less frequently. Expression can only be achieved in a bacterial strain carrying the gene for the T7 RNA polymerase. The most common cell strain to use with a T7 promoter system is BL21(DE3) (competent cells are chemically competent cells used for high-level protein expression with T7 RNA polymerase-based expression systems). The BL21(DE3) contains the T7 RNA polymerase gene, under the control of the lacUV5 promoter, integrated into the chromosome. IPTG is used to induce the expression of recombinant proteins cloned into vectors downstream of a T7 RNA promoter and transformed into the BL21(DE3) cells.
The T7 promoter is a BioBrick part BBa_1712074
C-terminal His-tag. In our protein expression construct, there is an amino acid motif that consists of six histidine (His) residues fused to the C-terminus of RiAFP. Histidine tags are widely used because they are small and rarely interfere with the function, activity, or structure of target proteins. The polyhistidine-tag is to be used to detect the secreted protein via anti-polyhistidine-tag antibodies or alternatively by in-gel staining (SDS-PAGE) with fluorescent probes bearing metal ions.
HlyA-signal peptide. The HlyA is a signal peptide found in the C-terminal signal sequence of alpha-hemolysin (HlyA). It is used to target RiAFP for secretion via the Type I secretion pathway of gram-negative bacteria. Fusion of the HlyA signal peptide to RiAFP results in transport of the protein from the cytoplasm to the extracellular medium in a single step.
The HlyA signal peptide is BioBrick pat BBa_K208006
Terminator. There are several E. coli transcriptional terminators available via the Registry. The most commonly used type of terminator is a forward terminator. When placed downstream of a genetic part that is transcribed, a forward transcriptional terminator will cause transcription to abort. We use part BBa_B0015, a double terminator (B0010-B0012). BBa_B0015. This terminator has an Average Forward Termination Efficiency of 98.4%.
Our PlantiFreeze device is a dual plasmid system in BL21 DE3 E coli.

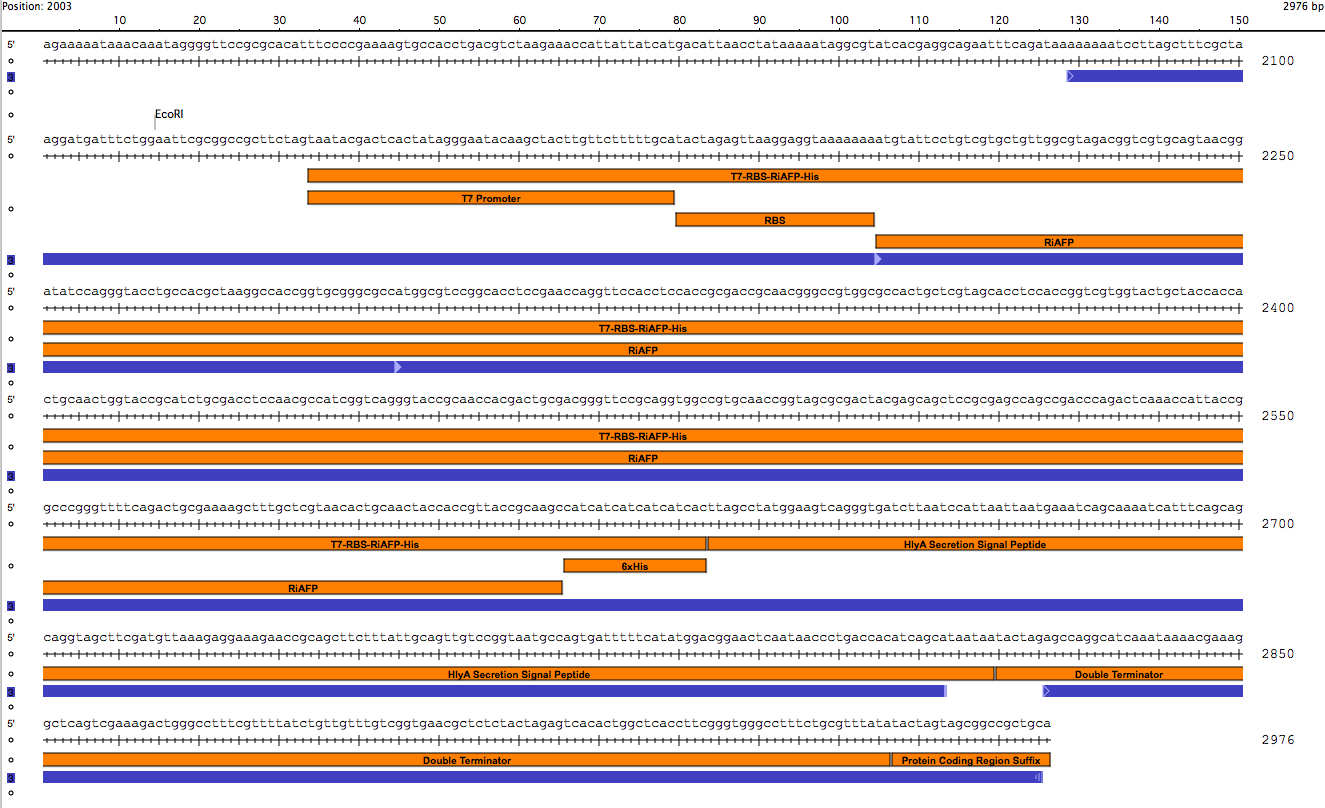
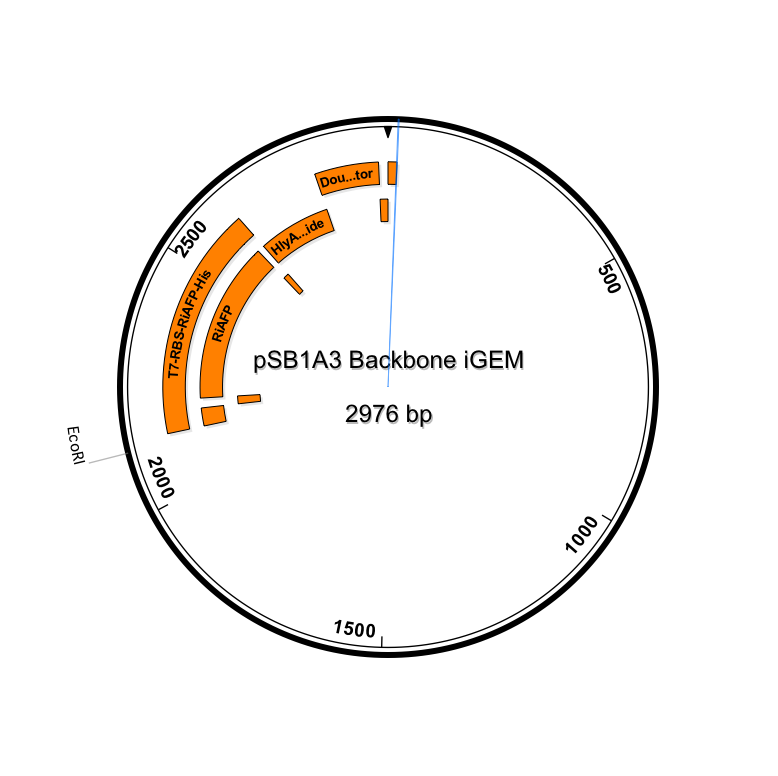
Sequence outlined in SeqBuilder (click to enlarge). The image on the left is the single-strand DNA sequence of the part. The image on the right is the plasmid map.
 "
"