Team:CIDEB-UANL Mexico/project aroma
From 2014hs.igem.org
DiegoValadez (Talk | contribs) |
|||
| (6 intermediate revisions not shown) | |||
| Line 25: | Line 25: | ||
<img width=131 height=127 src="https://static.igem.org/mediawiki/2014hs/5/5c/HandsomeAroma.jpg" align=right hspace=12/> | <img width=131 height=127 src="https://static.igem.org/mediawiki/2014hs/5/5c/HandsomeAroma.jpg" align=right hspace=12/> | ||
| + | <br> | ||
<p align="justify">Since the beginning of iGEM project, the use of fluorescent reporters has been used in each one of the proposed projects in previous years, trying to test the theoretical presence of other proteins in <i>E. coli.</i> For our iGEM 2014 project, this module proposed to promote the usage of aroma reporters, instead of fluorescent ones.</p> | <p align="justify">Since the beginning of iGEM project, the use of fluorescent reporters has been used in each one of the proposed projects in previous years, trying to test the theoretical presence of other proteins in <i>E. coli.</i> For our iGEM 2014 project, this module proposed to promote the usage of aroma reporters, instead of fluorescent ones.</p> | ||
| - | + | <br> | |
<p><b><h2>Description</h2></b></p> | <p><b><h2>Description</h2></b></p> | ||
| - | <p>In the module, WinterGreen | + | <p>In the module, WinterGreen is the most important part. Normally, it catalyzes the conversion of salicylic acid into methyl salicylate. The protein is excreted by the bacteria and when salicylic acid is added to the medium, a chemical reaction takes place and produces methyl salicylate, which is responsible for the wintergreen odor.</p> |
<p>But in this module, it will be regulated by temperature with the use of the RNA thermometer. When adding salicylic acid to the bacteria in a 32° Celsius environment, the production of the WinterGreen protein will begin.</p><br> | <p>But in this module, it will be regulated by temperature with the use of the RNA thermometer. When adding salicylic acid to the bacteria in a 32° Celsius environment, the production of the WinterGreen protein will begin.</p><br> | ||
| Line 44: | Line 45: | ||
<p><b>Figure 2.</b> The device originally thought, and the 2 modules derived from it.</p></center><br> | <p><b>Figure 2.</b> The device originally thought, and the 2 modules derived from it.</p></center><br> | ||
| - | <p>This device is composed by the following parts (see <b>figure 3</b>): | + | <p>This device is composed by the following parts (see <b>figure 3</b>): a constitutive promoter (1), a RNA thermometer (3); used to regulate the WinterGreen-odor protein production through temperature, a Wintergreen-odor enzyme generator, used to allow the production of methyl salicylate, induced by salicylic acid (2); and a terminator (4). All of these parts are ligated by an 8-bp scar (TACTAGAG).</p> |
<center><img width=400 height=170 src="https://static.igem.org/mediawiki/2014hs/5/56/AromaFigure1Aa.png" align=center hspace=12/> | <center><img width=400 height=170 src="https://static.igem.org/mediawiki/2014hs/5/56/AromaFigure1Aa.png" align=center hspace=12/> | ||
| Line 101: | Line 102: | ||
</span></b></p> | </span></b></p> | ||
</td> | </td> | ||
| - | <td width=170 valign=top style='width: | + | <td width=170 valign=top style='width:127.35pt;border-top:none;border-left: none;border-bottom:solid #C2D69B 1.0pt;border-right:solid #C2D69B 1.0pt; mso-border-top-alt:solid #C2D69B .5pt;mso-border-left-alt:solid #C2D69B .5pt; mso-border-alt:solid #C2D69B .5pt;background:#EAF1DD;padding:0cm 5.4pt 0cm 5.4pt; height:33.0pt'> |
<p class=MsoNormal style='margin-bottom:0cm;margin-bottom:.0001pt;line-height: normal'> | <p class=MsoNormal style='margin-bottom:0cm;margin-bottom:.0001pt;line-height: normal'> | ||
| Line 117: | Line 118: | ||
<p class=MsoNormal style='margin-bottom:0cm;margin-bottom:.0001pt;line-height: normal'> | <p class=MsoNormal style='margin-bottom:0cm;margin-bottom:.0001pt;line-height: normal'> | ||
| - | <span style='font-size:12.0pt;mso-ansi-language:EN-US'>Temperature-sensing bacteria that | + | <span style='font-size:12.0pt;mso-ansi-language:EN-US'>Temperature-sensing bacteria that changes color at different temperatures; as a temperature reporter system inlarge-scale fermentations, or as a temperature-inducible protein production system.<o:p></o:p> |
</span></p> | </span></p> | ||
| Line 131: | Line 132: | ||
<p class=MsoNormal style='margin-bottom:0cm;margin-bottom:.0001pt;line-height: normal'><b><u> | <p class=MsoNormal style='margin-bottom:0cm;margin-bottom:.0001pt;line-height: normal'><b><u> | ||
| - | <span style='font-size:12.0pt;color:#4F81BD;mso-ansi-language: EN-US'><a href="https:// | + | <span style='font-size:12.0pt;color:#4F81BD;mso-ansi-language: EN-US'><a href="https://2009.igem.org/Team:VictoriaBC" target="_blank">Victoria BC 2009</a> |
</span></u></b> | </span></u></b> | ||
| Line 141: | Line 142: | ||
<span style='font-size:12.0pt;mso-ansi-language:EN-US'><o:p> </o:p> | <span style='font-size:12.0pt;mso-ansi-language:EN-US'><o:p> </o:p> | ||
</span></b></p> | </span></b></p> | ||
| + | </td> | ||
| + | <td width=170 valign=top style='width:127.35pt;border-top:none;border-left: none;border-bottom:solid #C2D69B 1.0pt;border-right:solid #C2D69B 1.0pt; mso-border-top-alt:solid #C2D69B .5pt;mso-border-left-alt:solid #C2D69B .5pt; mso-border-alt:solid #C2D69B .5pt;padding:0cm 5.4pt 0cm 5.4pt'> | ||
| + | |||
| + | <p class=MsoNormal style='margin-bottom:0cm;margin-bottom:.0001pt;line-height: normal'> | ||
| + | |||
| + | <span style='font-size:12.0pt;mso-ansi-language:EN-US'>RNA themometer<o:p></o:p> | ||
| + | </span></p> | ||
| + | |||
| + | <p class=MsoNormal style='margin-bottom:0cm;margin-bottom:.0001pt;line-height: normal'> | ||
| + | |||
| + | <span style='font-size:12.0pt;mso-ansi-language:EN-US'><o:p> </o:p> | ||
| + | </span></p> | ||
</td> | </td> | ||
<td width=387 valign=top style='width:290.6pt;border-top:none;border-left: none;border-bottom:solid #C2D69B 1.0pt;border-right:solid #C2D69B 1.0pt; mso-border-top-alt:solid #C2D69B .5pt;mso-border-left-alt:solid #C2D69B .5pt; mso-border-alt:solid #C2D69B .5pt;padding:0cm 5.4pt 0cm 5.4pt'> | <td width=387 valign=top style='width:290.6pt;border-top:none;border-left: none;border-bottom:solid #C2D69B 1.0pt;border-right:solid #C2D69B 1.0pt; mso-border-top-alt:solid #C2D69B .5pt;mso-border-left-alt:solid #C2D69B .5pt; mso-border-alt:solid #C2D69B .5pt;padding:0cm 5.4pt 0cm 5.4pt'> | ||
| Line 146: | Line 159: | ||
<p class=MsoNormal style='margin-bottom:0cm;margin-bottom:.0001pt;line-height: normal'> | <p class=MsoNormal style='margin-bottom:0cm;margin-bottom:.0001pt;line-height: normal'> | ||
| - | <span style='font-size:12.0pt;mso-ansi-language:EN-US'> | + | <span style='font-size:12.0pt;mso-ansi-language:EN-US'>NAND logic gate using the ribo-key/ribo-lock system designed by <a href="https://2006.igem.org/wiki/index.php/Berkeley">Berkeley 2006</a> team , producing RFP except when the cells are grown in the presence of both arabinose and IPTG, also coupling fluorescent outputs with the ribo-thermometers made by TUDelft 2008 team.<o:p></o:p> |
| + | </span></p> | ||
| + | |||
| + | <p class=MsoNormal style='margin-bottom:0cm;margin-bottom:.0001pt;line-height: normal'> | ||
| + | |||
| + | <span style='font-size:12.0pt;mso-ansi-language:EN-US'><o:p> </o:p> | ||
| + | </span></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr style='mso-yfti-irow:2'> | ||
| + | <td width=170 valign=top style='width:127.35pt;border:solid #C2D69B 1.0pt; border-top:none;mso-border-top-alt:solid #C2D69B .5pt;mso-border-alt:solid #C2D69B .5pt; padding:0cm 5.4pt 0cm 5.4pt'> | ||
| + | |||
| + | <p class=MsoNormal style='margin-bottom:0cm;margin-bottom:.0001pt;line-height: normal'><b><u> | ||
| + | |||
| + | <span style='font-size:12.0pt;color:#4F81BD;mso-ansi-language: EN-US'><a href="https://2013hs.igem.org/Team:CIDEB-UANL_Mexico" target="_blank">CIDEB UANL 2013</a> | ||
| + | </span></u></b> | ||
| + | |||
| + | <span style='font-size:6.0pt;mso-bidi-font-size:12.0pt;mso-ansi-language:EN-US; mso-bidi-font-weight:bold'>:<b> <o:p></o:p></b> | ||
| + | </span></p> | ||
| + | |||
| + | <p class=MsoNormal style='margin-bottom:0cm;margin-bottom:.0001pt;line-height: normal'><b> | ||
| + | |||
| + | <span style='font-size:12.0pt;mso-ansi-language:EN-US'><o:p> </o:p> | ||
| + | </span></b></p> | ||
| + | </td> | ||
| + | <td width=170 valign=top style='width:127.35pt;border-top:none;border-left: none;border-bottom:solid #C2D69B 1.0pt;border-right:solid #C2D69B 1.0pt; mso-border-top-alt:solid #C2D69B .5pt;mso-border-left-alt:solid #C2D69B .5pt; mso-border-alt:solid #C2D69B .5pt;padding:0cm 5.4pt 0cm 5.4pt'> | ||
| + | |||
| + | <p class=MsoNormal style='margin-bottom:0cm;margin-bottom:.0001pt;line-height: normal'> | ||
| + | |||
| + | <span style='font-size:12.0pt;mso-ansi-language:EN-US'>RNA themometer<o:p></o:p> | ||
| + | </span></p> | ||
| + | |||
| + | <p class=MsoNormal style='margin-bottom:0cm;margin-bottom:.0001pt;line-height: normal'> | ||
| + | |||
| + | <span style='font-size:12.0pt;mso-ansi-language:EN-US'><o:p> </o:p> | ||
| + | </span></p> | ||
| + | </td> | ||
| + | <td width=387 valign=top style='width:290.6pt;border-top:none;border-left: none;border-bottom:solid #C2D69B 1.0pt;border-right:solid #C2D69B 1.0pt; mso-border-top-alt:solid #C2D69B .5pt;mso-border-left-alt:solid #C2D69B .5pt; mso-border-alt:solid #C2D69B .5pt;padding:0cm 5.4pt 0cm 5.4pt'> | ||
| + | |||
| + | <p class=MsoNormal style='margin-bottom:0cm;margin-bottom:.0001pt;line-height: normal'> | ||
| + | |||
| + | <span style='font-size:12.0pt;mso-ansi-language:EN-US'>Production of Vip3ca3, which acts as a pesticide protein, regulated by specific temperatures in order to avoid overproduction and it will show activity against target organisms Coleoptera and Lepidoptera..<o:p></o:p> | ||
</span></p> | </span></p> | ||
| Line 160: | Line 214: | ||
<p class=MsoNormal style='margin-bottom:0cm;margin-bottom:.0001pt;line-height: normal'><b><u> | <p class=MsoNormal style='margin-bottom:0cm;margin-bottom:.0001pt;line-height: normal'><b><u> | ||
| - | <span lang=ES-MX style='font-size:12.0pt;color:#4F81BD'><a href="https:// | + | <span lang=ES-MX style='font-size:12.0pt;color:#4F81BD'><a href="https://2006.igem.org/wiki/index.php/MIT_2006" target="_blank">MIT 2006</a> |
</span></u></b> | </span></u></b> | ||
| Line 170: | Line 224: | ||
<span lang=ES-MX style='font-size:12.0pt'><o:p> </o:p> | <span lang=ES-MX style='font-size:12.0pt'><o:p> </o:p> | ||
</span></b></p> | </span></b></p> | ||
| + | </td> | ||
| + | <td width=170 valign=top style='width:127.35pt;border-top:none;border-left: none;border-bottom:solid #C2D69B 1.0pt;border-right:solid #C2D69B 1.0pt; mso-border-top-alt:solid #C2D69B .5pt;mso-border-left-alt:solid #C2D69B .5pt; mso-border-alt:solid #C2D69B .5pt;background:#EAF1DD;padding:0cm 5.4pt 0cm 5.4pt'> | ||
| + | |||
| + | <p class=MsoNormal style='margin-bottom:0cm;margin-bottom:.0001pt;line-height: normal'> | ||
| + | |||
| + | <span style='font-size:12.0pt;mso-ansi-language:EN-US'>BSMT1<o:p></o:p> | ||
| + | </span></p> | ||
</td> | </td> | ||
<td width=387 valign=top style='width:290.6pt;border-top:none;border-left: none;border-bottom:solid #C2D69B 1.0pt;border-right:solid #C2D69B 1.0pt; mso-border-top-alt:solid #C2D69B .5pt;mso-border-left-alt:solid #C2D69B .5pt; mso-border-alt:solid #C2D69B .5pt;background:#EAF1DD;padding:0cm 5.4pt 0cm 5.4pt'> | <td width=387 valign=top style='width:290.6pt;border-top:none;border-left: none;border-bottom:solid #C2D69B 1.0pt;border-right:solid #C2D69B 1.0pt; mso-border-top-alt:solid #C2D69B .5pt;mso-border-left-alt:solid #C2D69B .5pt; mso-border-alt:solid #C2D69B .5pt;background:#EAF1DD;padding:0cm 5.4pt 0cm 5.4pt'> | ||
| Line 175: | Line 236: | ||
<p class=MsoNormal style='margin-bottom:0cm;margin-bottom:.0001pt;line-height: normal'> | <p class=MsoNormal style='margin-bottom:0cm;margin-bottom:.0001pt;line-height: normal'> | ||
| - | <span style='font-size:12.0pt;mso-ansi-language:EN-US'> | + | <span style='font-size:12.0pt;mso-ansi-language:EN-US'>This device produces methyl salicylate in the presence of salicylic acid. Methyl salicylate smells strongly of mint (wintergreen). Production of methyl salicylate was verified both by scent and by gas chromatography: E. coli with no WGD did not produce methyl salicylate when SA was added to the medium, while E. coli with the WGD did produce methyl salicylate when SA was added to the medium..<o:p></o:p> |
</span></p> | </span></p> | ||
</td> | </td> | ||
| Line 181: | Line 242: | ||
</table> | </table> | ||
</div> | </div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<p><b><h2>Project Zoom in</h2></b></p> | <p><b><h2>Project Zoom in</h2></b></p> | ||
Latest revision as of 03:27, 21 June 2014
Aroma Module

Since the beginning of iGEM project, the use of fluorescent reporters has been used in each one of the proposed projects in previous years, trying to test the theoretical presence of other proteins in E. coli. For our iGEM 2014 project, this module proposed to promote the usage of aroma reporters, instead of fluorescent ones.
Description
In the module, WinterGreen is the most important part. Normally, it catalyzes the conversion of salicylic acid into methyl salicylate. The protein is excreted by the bacteria and when salicylic acid is added to the medium, a chemical reaction takes place and produces methyl salicylate, which is responsible for the wintergreen odor.
But in this module, it will be regulated by temperature with the use of the RNA thermometer. When adding salicylic acid to the bacteria in a 32° Celsius environment, the production of the WinterGreen protein will begin.

Figure 1. Production of Wintergreen Odor
Device
Originally, WinterGreen was thought to be the reporter of the Capture module, but in order to prove the function of this gene and also the one in charge of the capture of sodium ions (NhaS), we decided to separate the full device into the actual modules of Capture and Aroma, as seen in the figure below.
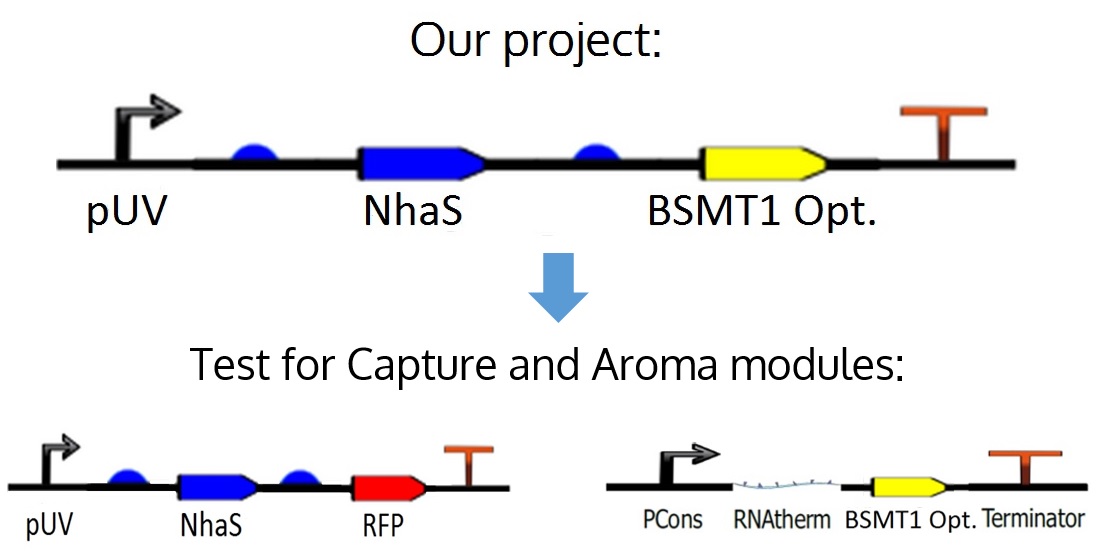
Figure 2. The device originally thought, and the 2 modules derived from it.
This device is composed by the following parts (see figure 3): a constitutive promoter (1), a RNA thermometer (3); used to regulate the WinterGreen-odor protein production through temperature, a Wintergreen-odor enzyme generator, used to allow the production of methyl salicylate, induced by salicylic acid (2); and a terminator (4). All of these parts are ligated by an 8-bp scar (TACTAGAG).

Figure 3. Aroma Module
These 4 genetic parts form the Aroma device of the project. The full device's length is 1,251bp (including restriction sites).
Parts of the module
| IMAGE | CODE | DESCRIPTION |
|
|
| In the specific case of our aroma module, it will help the bacteria to continuously transcribe the WinterGreen gene in order to allow the bacteria to continuously produce the aroma. This promoter has a length of 35bp. |
|
| A RNA thermometer, used for temperature post-transcriptional regulation (thermo sensor), and is designed to initiate transcription around 32°C. | |
|
|
| Produces a transferase to convert salicylic acid into methyl salicylate (WinterGreen odor). The wintergreen odor generator requires of 2mM of salicylic acid to produce methyl salicylate. BSMT1 (BBa_J45004), WinterGreen Odor Generator original name, was created by MIT 2006. This year, the team optimized the sequence for Escherichia coli. The new biobrick has a length of 1,074bp. |
|
|
| Part made of 6bp, responsible for transcription stop. The terminator stops the production of methyl salicylate. |
Justification
Different to fluorescent reporters, this module was made in order to (in the future) perform as an aroma reporter and also to test the correct function of the bacteria, for its future usage as a new reporter and functional part (CDS). It is desired to use this part in the project to replace the red fluorescent protein (RFP) in the Capture module. But it was preferable to test it apart to demonstrate its effectiveness. Similarly, this piece is also helpful when the biofilter is assembled, because when performing the filtration by silica, WinterGreen can demonstrate the presence of bacteria in the beads.
The team added the RNA thermometer in the device for regulating the production of the aroma. Another reason for selecting the RNA thermometer as a regulator was to continue the CIDEB UANL 2013 work with it.
Other teams that used RNA thermometer and BSMT1
|
Team |
Part Used |
Use |
|
RNA themometer
|
Temperature-sensing bacteria that changes color at different temperatures; as a temperature reporter system inlarge-scale fermentations, or as a temperature-inducible protein production system.
|
|
|
|
RNA themometer
|
NAND logic gate using the ribo-key/ribo-lock system designed by Berkeley 2006 team , producing RFP except when the cells are grown in the presence of both arabinose and IPTG, also coupling fluorescent outputs with the ribo-thermometers made by TUDelft 2008 team.
|
|
|
RNA themometer
|
Production of Vip3ca3, which acts as a pesticide protein, regulated by specific temperatures in order to avoid overproduction and it will show activity against target organisms Coleoptera and Lepidoptera..
|
|
MIT 2006
:
|
BSMT1 |
This device produces methyl salicylate in the presence of salicylic acid. Methyl salicylate smells strongly of mint (wintergreen). Production of methyl salicylate was verified both by scent and by gas chromatography: E. coli with no WGD did not produce methyl salicylate when SA was added to the medium, while E. coli with the WGD did produce methyl salicylate when SA was added to the medium.. |
Project Zoom in
Bibliography/References
● Huang, H. (2006, August 30). Part:BBa_B1002. Retrieved August 30, 2014, from http://parts.igem.org/wiki/index.php?title=Part:BBa_B1002.
● iGEM2006_Berkeley. (2006). Part:BBa_J23100. Retrieved August 30, 2014, from http://parts.igem.org/Part:BBa_J23100.
● iGEM2006_MIT. (2006). Part:BBa_J45004. Retrieved August 30, 2014, from http://parts.igem.org/Part:BBa_J45004.
● iGEM CIDEB Team. (2013). iGEM CIDEB UANL 2013. Retrieved on March 31th, 2014. https://2013hs.igem.org/Team:CIDEB-UANL_Mexico/Project.
● iGEM08_TUDelft. (2008). Part:BBa_K115017. Retrieved August 30, 2014, from http://parts.igem.org/Part:BBa_K115017.
● MIT IGEM Team. (2006). MIT 2006. Retrieved on March 31th, 2014, from: https://2006.igem.org/wiki/index.php/MIT_2006.
● TUDelft iGEM Team. (2008). TUDelft 2008. Retrieved on March 31th, 2014, from: https://2008.igem.org/Team:TUDelft.
● VictoriaBC. (2009). VictoriaBC 2009. Retrieved on March 31th, 2014, from: https://2009.igem.org/Team:VictoriaBC.
● Zubieta, Chole et al. (2003). Structural Basis for Substrate Recognition in the Salicylic Acid Carboxyl Methyltransferase Family. Manuscript submitted for publication. Retrieved from www.plantcell.org.
 "
"


















