Team:CSWProteens/project/Methods
From 2014hs.igem.org
| (9 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
<html xmlns="http://www.w3.org/1999/xhtml"> | <html xmlns="http://www.w3.org/1999/xhtml"> | ||
| - | + | <style> | |
| + | #contentSub, #footer-box, #catlinks, #search-controls, #p-logo, .printfooter, .firstHeading,.visualClear {display: none;} /*-- hides default wiki settings --*/ | ||
| + | </style> | ||
<head> | <head> | ||
<meta content="en-ca" http-equiv="Content-Language" /> | <meta content="en-ca" http-equiv="Content-Language" /> | ||
| Line 127: | Line 129: | ||
<meta content="revealTrans(Duration=3.0,Transition=2)" http-equiv="Site-Enter" /> | <meta content="revealTrans(Duration=3.0,Transition=2)" http-equiv="Site-Enter" /> | ||
</head> | </head> | ||
| - | <center><a href="https://2014hs.igem.org/Team:CSWProteens/project"><img alt="" class="auto-style11" height="200" src="https://static.igem.org/mediawiki/2014hs/c/cd/Plantifreeze.gif" width="200" /><img src="https://static.igem.org/mediawiki/2014hs/ | + | <center><a href="https://2014hs.igem.org/Team:CSWProteens/project"><img alt="" class="auto-style11" height="200" src="https://static.igem.org/mediawiki/2014hs/c/cd/Plantifreeze.gif" width="200" /><img src="https://static.igem.org/mediawiki/2014hs/7/7e/CSW_Banner.png" width="500"> |
</a> | </a> | ||
| - | <a href="https://2014hs.igem.org/Main_Page"><img alt="" class="auto-style5" height="200" src="https://static.igem.org/mediawiki/2014hs/ | + | <a href="https://2014hs.igem.org/Main_Page"><img alt="" class="auto-style5" height="200" src="https://static.igem.org/mediawiki/2014hs/2/26/Igem_grpyon.png" width="200" /></a> </p></center> |
<body style=" color: #FFFFFF;background-color: #000000; | <body style=" color: #FFFFFF;background-color: #000000; | ||
| Line 169: | Line 171: | ||
<a href="https://2014hs.igem.org/Team:CSWProteens/humanpractices"><b>Outreach</b></a> | <a href="https://2014hs.igem.org/Team:CSWProteens/humanpractices"><b>Outreach</b></a> | ||
<ul> | <ul> | ||
| - | + | ||
</ul> | </ul> | ||
</li> | </li> | ||
| Line 180: | Line 182: | ||
</ul> | </ul> | ||
</ul> | </ul> | ||
| + | <center><p class="auto-style4">M E T H O D S</center><p> | ||
<br/><p class="auto-style1"> | <br/><p class="auto-style1"> | ||
This section of our team Wiki gives an overall summary of the various methods we used to construct our PlantiFreeze device. It is important to note that the device is based on a dual plasmid design: one plasmid controlling expression of the recombinant protein, RiAFP, and the other plasmid controlling secretion of the protein. These procedures reflect the revised construction strategy implemented following our discovery that the wrong DNA sequence was originally ordered for synthesis by IDT. | This section of our team Wiki gives an overall summary of the various methods we used to construct our PlantiFreeze device. It is important to note that the device is based on a dual plasmid design: one plasmid controlling expression of the recombinant protein, RiAFP, and the other plasmid controlling secretion of the protein. These procedures reflect the revised construction strategy implemented following our discovery that the wrong DNA sequence was originally ordered for synthesis by IDT. | ||
| Line 193: | Line 196: | ||
<p><p class="auto-style1"> | <p><p class="auto-style1"> | ||
Overnight cultures were then used to seed larger 250 ml flasks (50 ml media volume) at an initial optical density (OD600) of 0.05 at time 0 h. 0.1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Sigma Aldrich, Inc. St. Louis, MO) was added to each flask. Flasks were removed at 24, and 48 h and media and lyzed cells were analyzed for RiAFP with the use of InVision™ His-tag In-gel Stain (Life Technologies, Carlsbad, CA), a fluorescent stain that is specially formulated for fast, sensitive, and specific detection of His-tagged fusion proteins. Electrophoresis of the sample is run on a polyacrylamide gel (1.0-mm thick NuPAGE® Novex Gel, Life Technologies, Carlsbad, CA). The staining is capable of detecting ~0.5 picomole of a 6X His-tagged fusion protein (e.g. 1 picomole of a 30 kDa protein is 30 ng). A His-tagged Protein Standard (BenchMark™ His-tagged Protein Standard, Life Technologies) is used as a positive control for the InVision™ His-tag In-gel Stain. After staining, His-tagged fusion protein bands are visualized with a UV transilluminator equipped with a camera. | Overnight cultures were then used to seed larger 250 ml flasks (50 ml media volume) at an initial optical density (OD600) of 0.05 at time 0 h. 0.1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Sigma Aldrich, Inc. St. Louis, MO) was added to each flask. Flasks were removed at 24, and 48 h and media and lyzed cells were analyzed for RiAFP with the use of InVision™ His-tag In-gel Stain (Life Technologies, Carlsbad, CA), a fluorescent stain that is specially formulated for fast, sensitive, and specific detection of His-tagged fusion proteins. Electrophoresis of the sample is run on a polyacrylamide gel (1.0-mm thick NuPAGE® Novex Gel, Life Technologies, Carlsbad, CA). The staining is capable of detecting ~0.5 picomole of a 6X His-tagged fusion protein (e.g. 1 picomole of a 30 kDa protein is 30 ng). A His-tagged Protein Standard (BenchMark™ His-tagged Protein Standard, Life Technologies) is used as a positive control for the InVision™ His-tag In-gel Stain. After staining, His-tagged fusion protein bands are visualized with a UV transilluminator equipped with a camera. | ||
| - | <p><center> | + | <p><center><a href="https://static.igem.org/mediawiki/2014hs/e/e4/Overall_methods1.jpg"> |
| - | <img src="https://static.igem.org/mediawiki/2014hs/ | + | <img src="https://static.igem.org/mediawiki/2014hs/e/e4/Overall_methods1.jpg" width="700"></a><br>Click to enlarge image above.</center> |
Latest revision as of 02:21, 21 June 2014



M E T H O D S
This section of our team Wiki gives an overall summary of the various methods we used to construct our PlantiFreeze device. It is important to note that the device is based on a dual plasmid design: one plasmid controlling expression of the recombinant protein, RiAFP, and the other plasmid controlling secretion of the protein. These procedures reflect the revised construction strategy implemented following our discovery that the wrong DNA sequence was originally ordered for synthesis by IDT.
Genetic parts were constructed in accordance with the BioBrick technical standard. The DNA sequence comprising the expression plasmid was synthesized as a gBlock™ DNA fragment by Integrated DNA Technologies (IDT, Coralville, IA). Using standard iGEM 3A assembly, the DNA fragment is ligated with the linearized pSB1A3 plasmid (see plasmid sequence and map). pSB1A3 is a BioBrick standard vector for assembly and expression of BioBrick genetic devices. All restriction enzymes and related reagents were donated by New England Biolabs, Ipswich, MA.
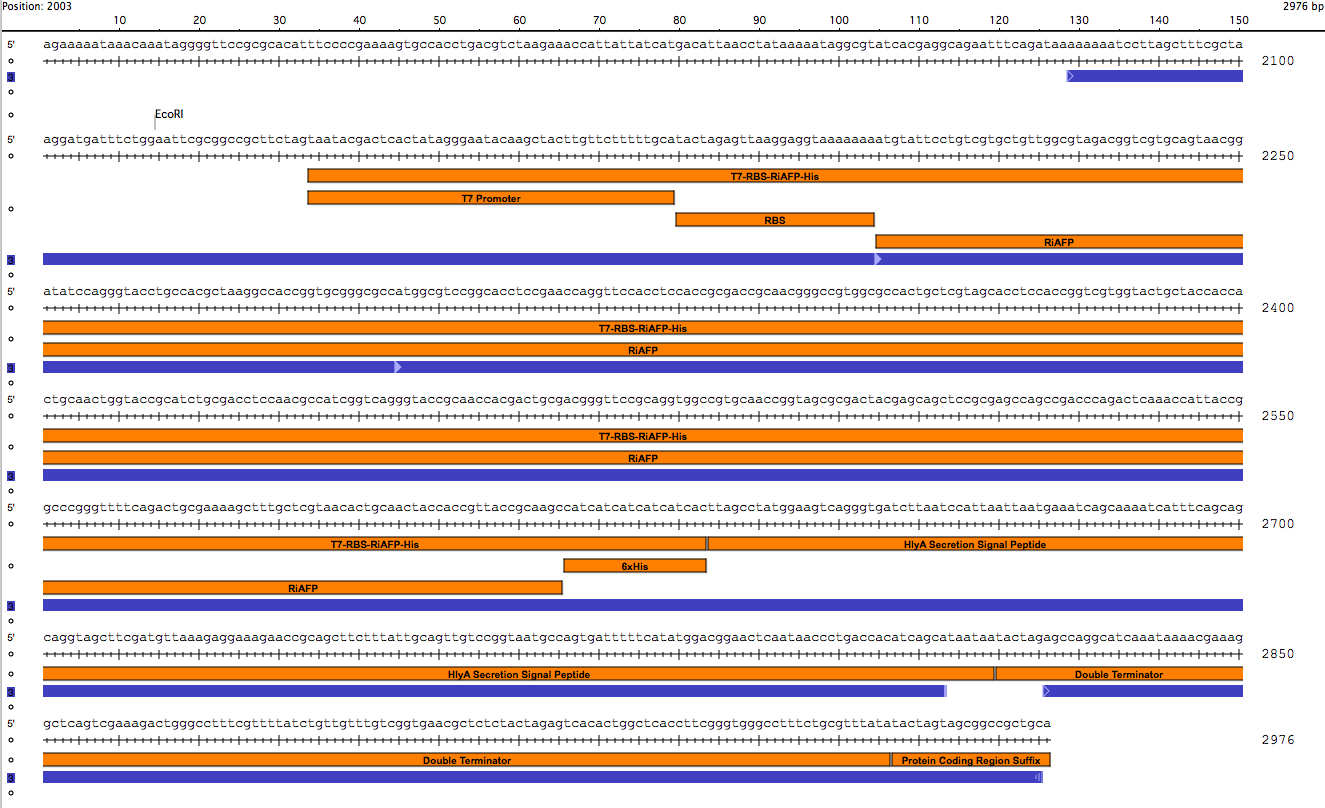
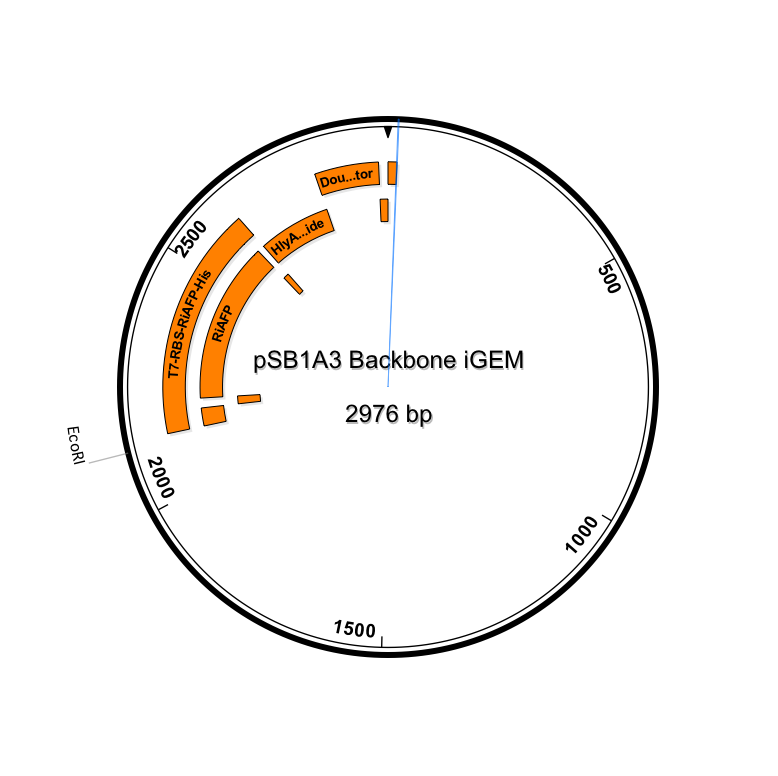
(click to enlarge)
The pSB1A3 plasmid with the DNA fragment insert is used to transform competent NEB 10 beta cells. The cells are plated on agar supplemented with ampicillin. After 16 hours incubation at 37° C, individual colonies are selected and grown up in 5ml of LB broth (with ampicillin) in 15 mL culture tubes.
Now that cell cultures of E. coli containing lots of copies of the expression plasmid have been grown up, Quiagen miniprep is used to access the expression plasmid DNA: lyse the cells, separate the DNA from the cellular material, and purify the sample to make sure you only have the plasmid DNA and not the E. coli's own genomic DNA. We used a Qiagen Miniprep Kit (Qiagen, Valencia, CA) and our mini-centrifuge with a max speed of 4400 rpm.
Thanks to the generosity of Asif Rahman, our team received the secretion plasmid, pLG575, from the Utah State University. Plasmid pLG575 includes the coding regions for proteins HlyB and HlyD and carries the gene for chloramphenicol resistance. The next step in the construction of our device is the cotransformation of competent BL21 DE3 cells (New England Biolabs) with the expression plasmid accessed by the miniprep and pLG575. The cells are plated on agar supplemented with ampicillin and chloramphenicol. After 16 hours incubation at 37° C, individual colonies are selected and grown up in 5ml of LB broth (with ampicillin and chloramphenical) in 15 mL culture tubes.
Overnight cultures were then used to seed larger 250 ml flasks (50 ml media volume) at an initial optical density (OD600) of 0.05 at time 0 h. 0.1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Sigma Aldrich, Inc. St. Louis, MO) was added to each flask. Flasks were removed at 24, and 48 h and media and lyzed cells were analyzed for RiAFP with the use of InVision™ His-tag In-gel Stain (Life Technologies, Carlsbad, CA), a fluorescent stain that is specially formulated for fast, sensitive, and specific detection of His-tagged fusion proteins. Electrophoresis of the sample is run on a polyacrylamide gel (1.0-mm thick NuPAGE® Novex Gel, Life Technologies, Carlsbad, CA). The staining is capable of detecting ~0.5 picomole of a 6X His-tagged fusion protein (e.g. 1 picomole of a 30 kDa protein is 30 ng). A His-tagged Protein Standard (BenchMark™ His-tagged Protein Standard, Life Technologies) is used as a positive control for the InVision™ His-tag In-gel Stain. After staining, His-tagged fusion protein bands are visualized with a UV transilluminator equipped with a camera.
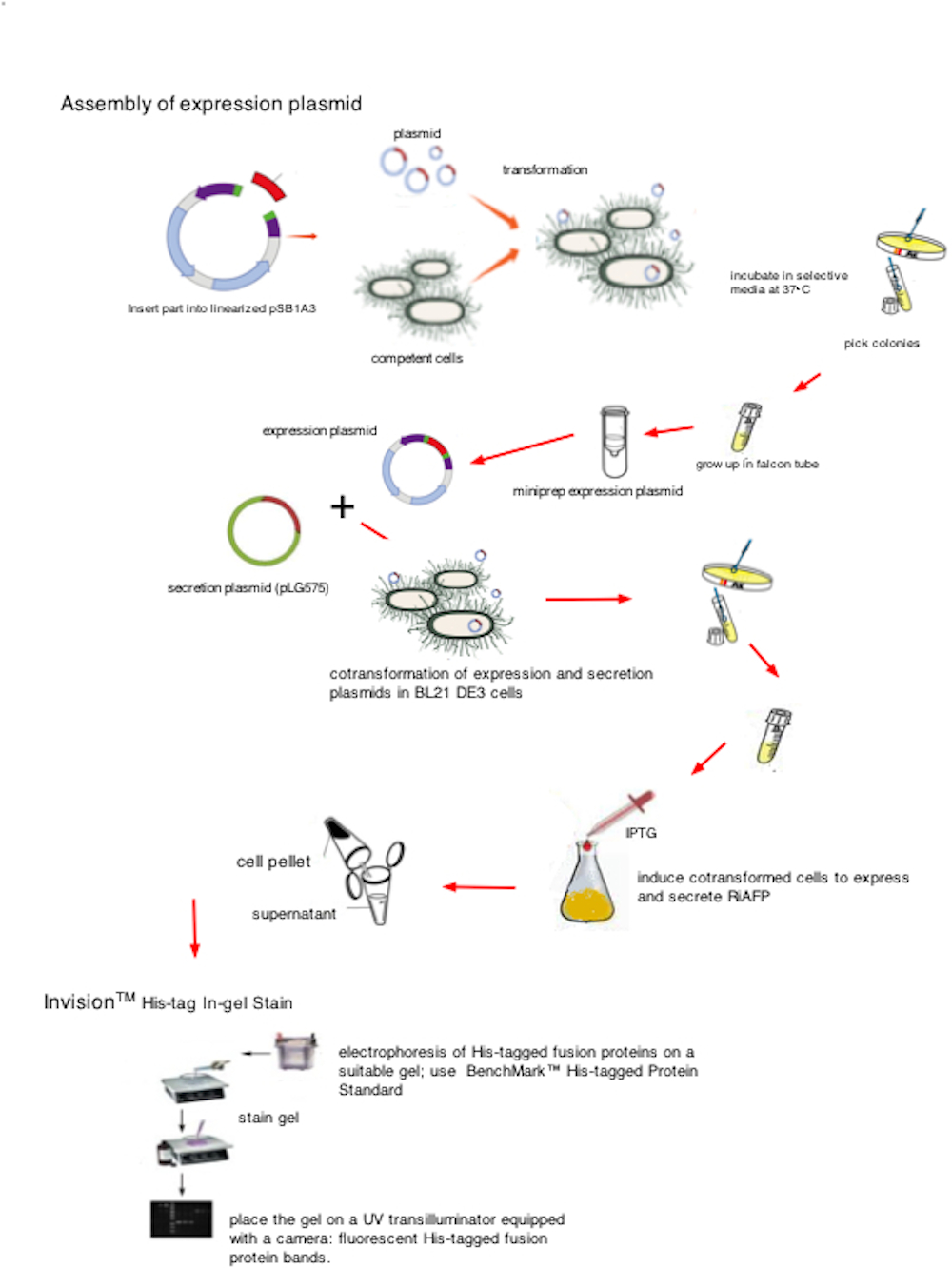
Click to enlarge image above.
 "
"